User login
The Burden of Skin Cancer in the Military Health System, 2017-2022
This retrospective observational study investigates skin cancer prevalence and care patterns within the Military Health System (MHS) from 2017 to 2022. Utilizing the MHS Management Analysis and Reporting Tool (most commonly called M2), we analyzed more than 5 million patient encounters and documented skin cancer prevalence in the MHS beneficiary population utilizing available demographic data. Notable findings included an increased prevalence of skin cancer in the military population compared with the civilian population, a substantial decline in direct care (DC) visits at military treatment facilities compared with civilian purchased care (PC) visits, and a decreased total number of visits during COVID-19 restrictions.
The Military Health System (MHS) is a worldwide health care delivery system that serves 9.6 million beneficiaries, including military service members, retirees, and their families.1 Its mission is 2-fold: provide a medically ready force, and provide a medical benefit in keeping with the service and sacrifice of active-duty personnel, military retirees, and their families. For fiscal year (FY) 2022, active-duty service members and their families comprised 16.7% and 19.9% of beneficiaries, respectively, while retired service members and their families comprised 27% and 32% of beneficiaries, respectively.
The MHS operates under the authority of the Department of Defense (DoD) and is supported by an annual budget of approximately $50 billion.1 Health care provision within the MHS is managed by TRICARE regional networks.2 Within these networks, MHS beneficiaries may receive health care in 2 categories: direct care (DC) and purchased care (PC). Direct care is rendered in military treatment facilities by military or civilian providers contracted by the DoD, and PC is administered by civilian providers at civilian health care facilities within the TRICARE network, which is comprised of individual providers, clinics, and hospitals that have agreed to accept TRICARE beneficiaries.1 Purchased care is fee-for-service and paid for by the MHS. Of note, the MHS differs from the Veterans Affairs health care system in that the MHS through DC and PC sees only active-duty service members, active-duty dependents, retirees, and retirees’ dependents (primarily spouses), whereas Veterans Affairs sees only veterans (not necessarily retirees) discharged from military service with compensable medical conditions or disabilities.
Skin cancer presents a notable concern for the US Military, as the risk for skin cancer is thought to be higher than in the general population.3,4 This elevated risk is attributed to numerous factors inherent to active-duty service, including time spent in tropical environments, increased exposure to UV radiation, time spent at high altitudes, and decreased rates of sun-protective behaviors.3 Although numerous studies have explored the mechanisms that contribute to service members’ increased skin cancer risk, there are few (if any) that discuss the burden of skin cancer on the MHS and where its beneficiaries receive their skin cancer care. This study evaluated the burden of skin cancer within the MHS, as demonstrated by the period prevalence of skin cancer among its beneficiaries and the number and distribution of patient visits for skin cancer across both DC and PC from 2017 to 2022.
Methods
Data Collection—This retrospective observational study was designed to describe trends in outpatient visits with a skin cancer diagnosis and annual prevalence of skin cancer types in the MHS. Data are from all MHS beneficiaries who were eligible or enrolled in the analysis year. Our data source was the MHS Management Analysis and Reporting Tool (most commonly called M2), a query tool that contains the current and most recent 5 full FYs of Defense Health Agency corporate health care data including aggregated FY and calendar-year counts of MHS beneficiaries from 2017 to 2022 using encounter and claims data tables from both DC and PC. Data in M2 are coded using a pseudo-person identification number, and queries performed for this study were limited to de-identified visit and patient counts.
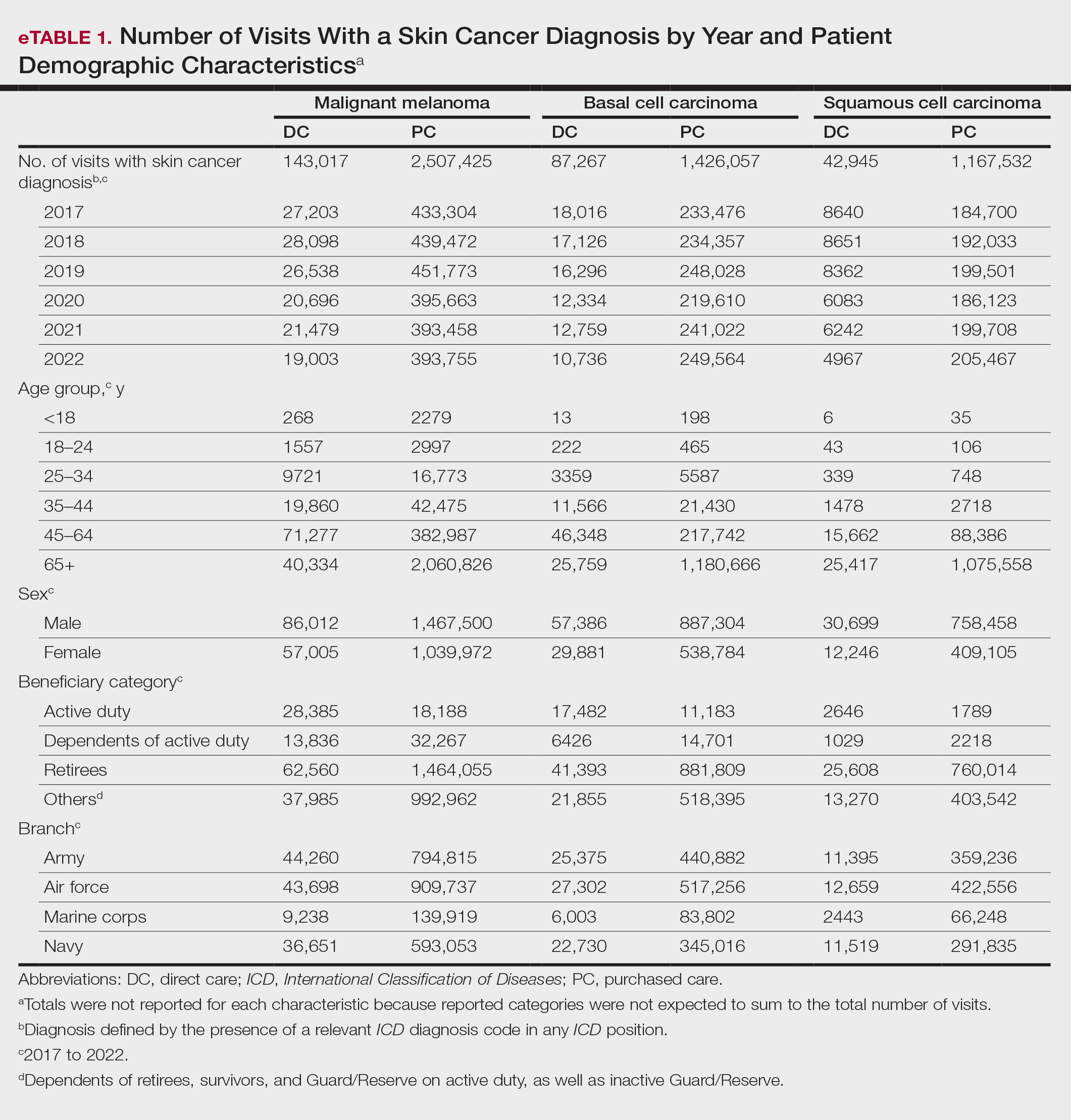
Skin cancer diagnoses were defined by relevant International Classification of Diseases, Tenth Revision, Clinical Modification (ICD-10-CM) codes recorded from outpatient visits in DC and PC. The M2 database was queried to find aggregate counts of visits and unique MHS beneficiaries with one or more diagnoses of a skin cancer type of interest (defined by relevant ICD-10-CM code) over the study period stratified by year and by patient demographic characteristics. Skin cancer types by ICD-10-CM code group included basal cell carcinoma (BCC), squamous cell carcinoma (SCC), malignant melanoma (MM), and other (including Merkel cell carcinoma and sebaceous carcinoma). Demographic strata included age, sex, military status (active duty, dependents of active duty, retired, or all others), sponsor military rank, and sponsor branch (army, air force, marine corps, or navy). Visit counts included diagnoses from any ICD position (for encounters that contained multiple ICD codes) to describe the total volume of care that addressed a diagnosed skin cancer. Counts of unique patients in prevalence analyses included relevant diagnoses in the primary ICD position only to increase the specificity of prevalence estimates.
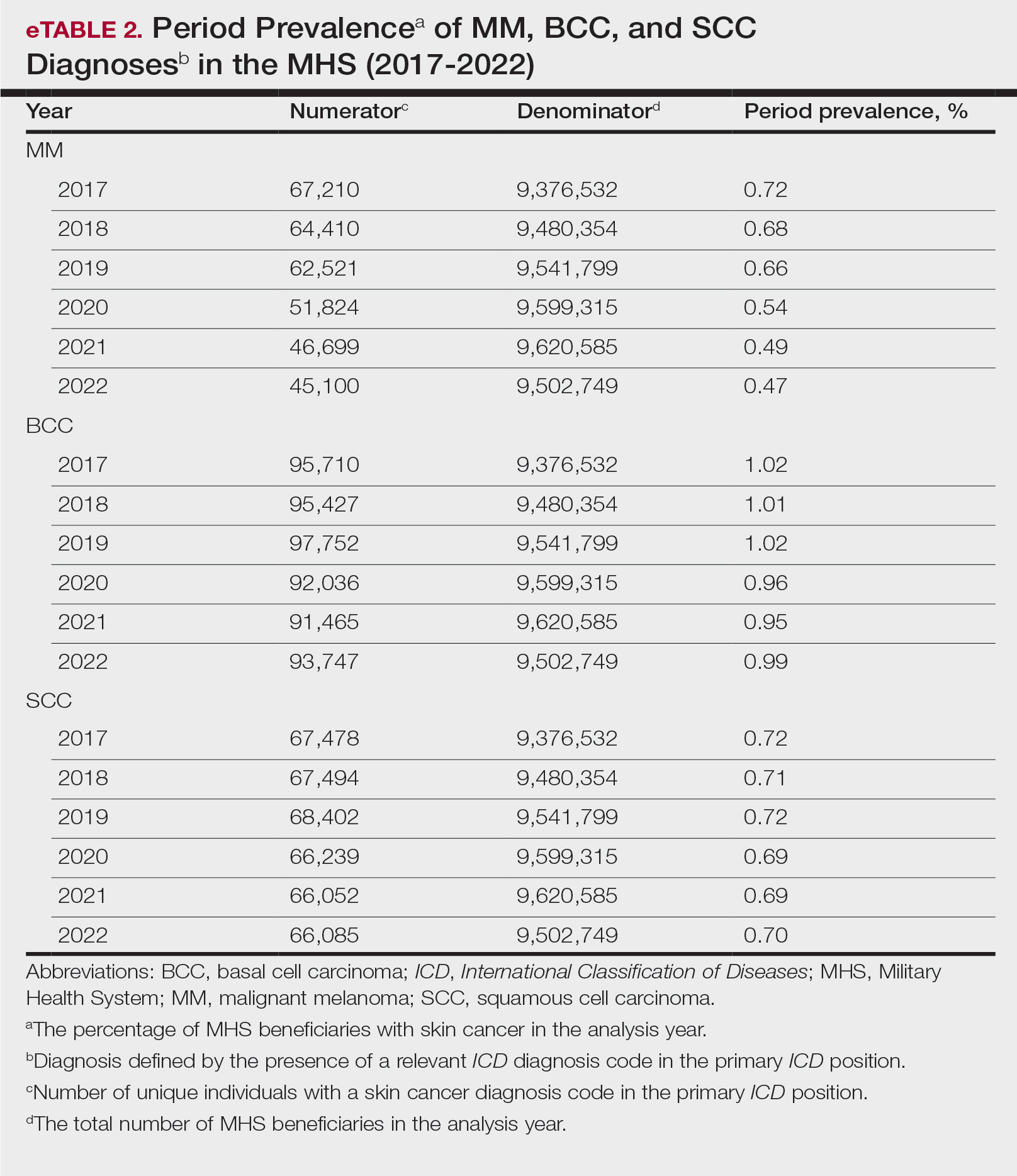
Data Analysis—Descriptive analyses included the total number of outpatient visits with a skin cancer diagnosis in DC and PC over the study period, with percentages of total visits by year and by demographic strata. Separate analyses estimated annual prevalences of skin cancer types in the MHS by study year and within 2022 by demographic strata. Numerators in prevalence analyses were defined as the number of unique individuals with one or more relevant ICD codes in the analysis year. Denominators were defined as the total number of MHS beneficiaries in the analysis year and resulting period prevalences reported. Observed prevalences were qualitatively described, and trends were compared with prevalences in nonmilitary populations reported in the literature.
Ethics—This study was conducted as part of a study using secondary analyses of de-identified data from the M2 database. The study was reviewed and approved by the Walter Reed National Military Medical Center institutional review board.
Results
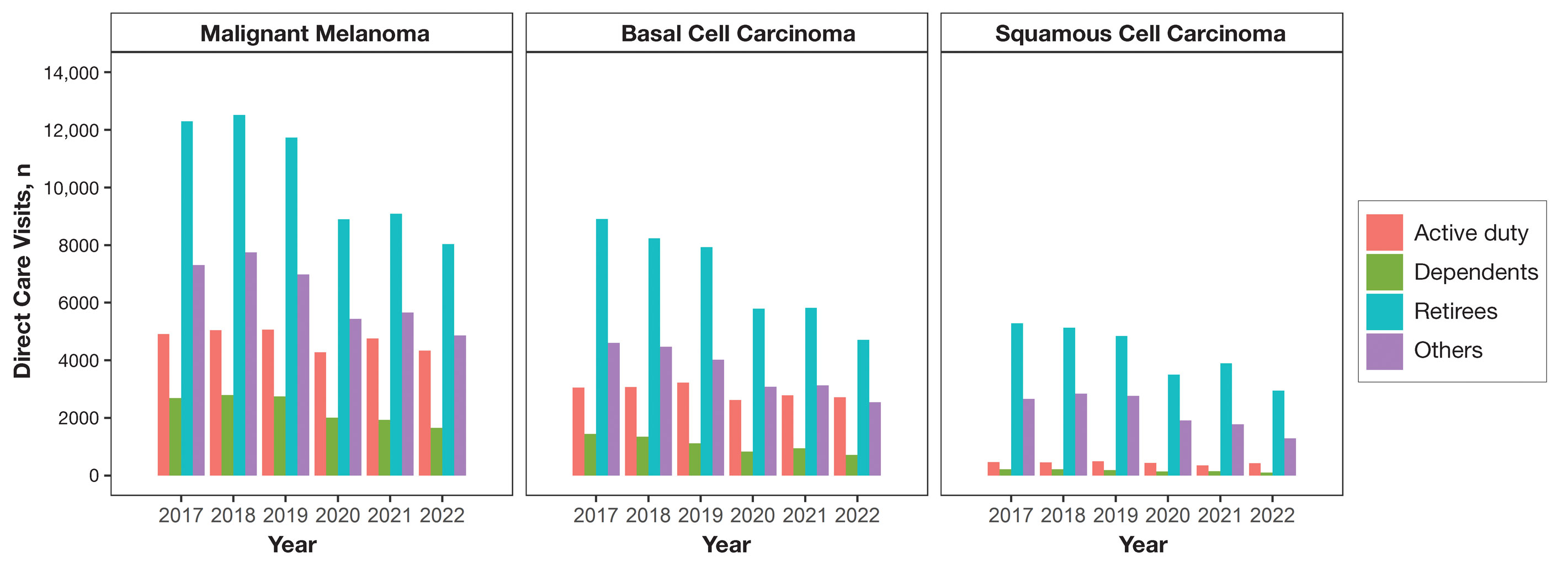
Encounter data were analyzed from a total of 5,374,348 visits between DC and PC over the study period for each cancer type of interest. Figures 1 and 2 show temporal trends in DC visits compared with PC visits in each beneficiary category. The percentage of total DC visits subsequently declined each year throughout the study period, with percentage decreases from 2017 to 2022 of 1.45% or 8200 fewer visits for MM, 3.41% or 7280 fewer visits for BCC, and 2.26% or 3673 fewer visits for SCC.
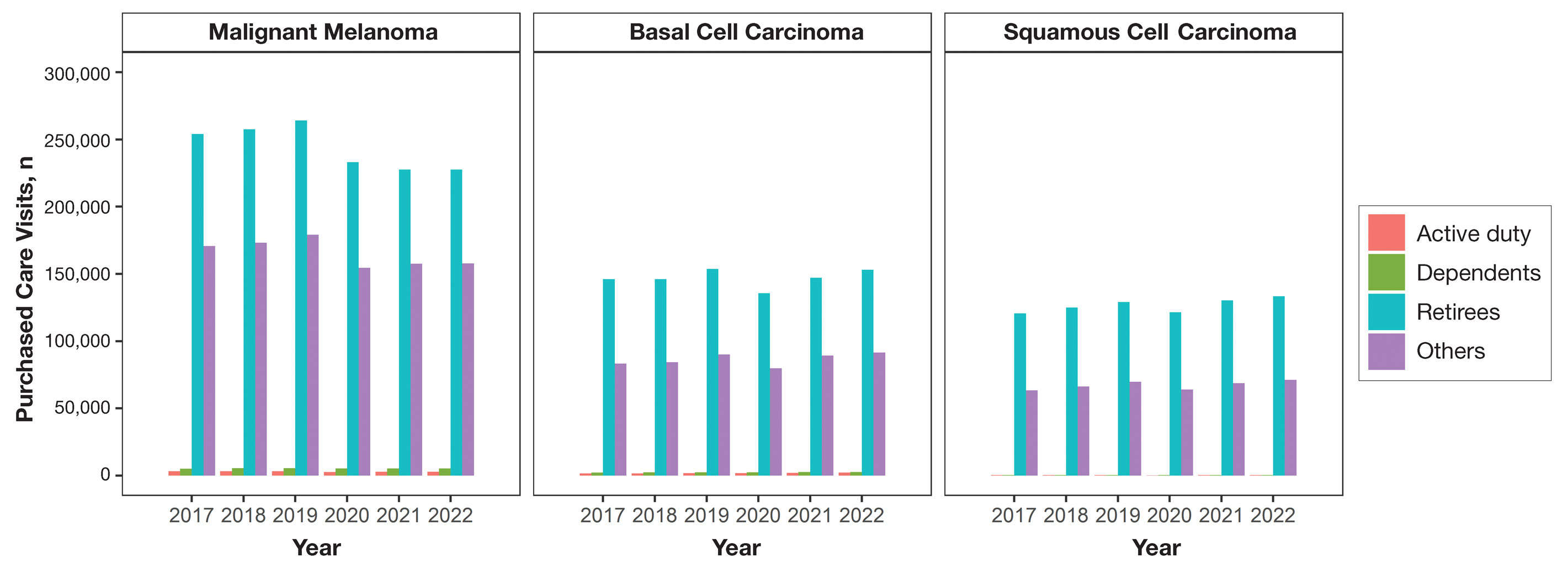
When stratified by beneficiary category, this trend remained consistent among dependents and retirees, with the most notable annual percentage decrease from 2019 to 2020. A higher proportion of younger adults and active-duty beneficiaries was seen in DC relative to PC, in which most visits were among retirees and others (primarily dependents of retirees, survivors, and Guard/Reserve on active duty, as well as inactive Guard/Reserve). No linear trends over time were apparent for active duty in DC and for dependents and retirees in PC. eTable 1 summarizes the demographic characteristics of MHS beneficiaries being seen in DC and PC over the study period for each cancer type of interest.
The Table shows the period prevalence of skin cancer diagnoses within the MHS beneficiary population from 2017 to 2022. These data were further analyzed by MM, BCC, and SCC (eTable 2) and demographics of interest for the year 2022. By beneficiary category, the period prevalence of MM was 0.08% in active duty, 0.06% in dependents, 0.48% in others, and 1.10% in retirees; the period prevalence of BCC was 0.12% in active duty, 0.07% in dependents, 0.91% in others, and 2.50% in retirees; and the period prevalence of SCC was 0.02% in active duty, 0.01% in dependents, 0.63% in others, and 1.87% in retirees. By sponsor branch, the period prevalence of MM was 0.35% in the army, 0.62% in the air force, 0.35% in the marine corps, and 0.65% in the navy; the period prevalence of BCC was 0.74% in the army, 1.30% in the air force, 0.74% in the marine corps, and 1.36% in the navy; and the period prevalence of SCC was 0.52% in the army, 0.92% in the air force, 0.51% in the marine corps, and 0.97% in the navy.

Comment
This study aimed to provide insight into the burden of skin cancer within the MHS beneficiary population and to identify temporal trends in where these beneficiaries receive their care. We examined patient encounter data from more than 9.6 million MHS beneficiaries.
The utilization of ICD codes from patient encounters to estimate the prevalence of nonmelanoma skin cancer (NMSC) has demonstrated a high positive predictive value. In one study, NMSC cases were confirmed in 96.5% of ICD code–identified patients.5 We presented an extensive collection of epidemiologic data on BCC and SCC, which posed unique challenges for tracking, as they are not reported to or monitored by cancer registries such as the Surveillance, Epidemiology, and End Results (SEER) Program.6
MHS Compared to the US Population—A study using the Global Burden of Disease 2019 database revealed an increasing trend in the incidence and prevalence of NMSC and melanoma since 1990. The same study found the period prevalence in 2019 of MM, SCC, and BCC in the general US population to be 0.13%, 0.31%, and 0.05%, respectively.7 In contrast, among MHS beneficiaries, we observed a higher prevalence in the same year, with figures of 0.66% for MM, 0.72% for SCC, and 1.02% for BCC. According to the SEER database, the period prevalence of MM within the general US population in 2020 was 0.4%.8 That same year, we identified a higher period prevalence of MM—0.54%—within the MHS beneficiary population. Specifically, within the MHS retiree population, the prevalence in 2022 was double that of the general MHS population, with a rate of 1.10%, underscoring the importance of skin cancer screening in older, at-risk adult populations. Prior studies similarly found increased rates of skin cancer within the military beneficiary population. Further studies are needed to compare age-adjusted rates in the MHS vs US population.9-11
COVID-19 Trends—Our data showed an overall decreasing prevalence of skin cancer in the MHS from 2019 to 2021. We suspect that the apparent decrease in skin cancer prevalence may be attributed to underdiagnosis from COVID-19 pandemic restrictions. During that time, many dermatology clinics at military treatment facilities underwent temporary closures, and some dermatologists were sent on nondermatologic utilization tours. Likewise, a US multi-institutional study described declining rates of new melanomas from 2020 to 2021, with an increased proportion of patient presentations with advanced melanoma, suggesting an underdiagnosis of melanoma cases during pandemic restrictions. That study also noted an increased rate of patient-identified melanomas and a decreased rate of provider-identified melanomas during that time.12 Contributing factors may include excess hospital demand, increased patient complexity and acute care needs, and long outpatient clinic backlogs during this time.13Financial Burden—Over our 5-year study period, there were 5,374,348 patient encounters addressing skin cancer, both in DC and PC (Figures 1 and 2; eTable 1). In 2016 to 2018, the average annual cost of treating skin cancer in the US civilian, noninstitutionalized population was $1243 for NMSC (BCC and SCC) and $2430 for melanoma.6 Using this metric, the estimated total cost of care rendered in the MHS in 2018 for NMSC and melanoma was $202,510,803 and $156,516,300, respectively.
Trends in DC vs PC—In the years examined, we found a notable decrease in the number of beneficiaries receiving treatment for MM, BCC, and SCC in DC. Simultaneously, there has been an increase in the number of beneficiaries receiving PC for BCC and SCC, though this trend was not apparent for MM.
Our data provided interesting insights into the percentage of PC compared with DC offered within the MHS. Importantly, our findings suggested that the majority of skin cancer in active-duty service members is managed with DC within the military treatment facility setting (61% DC management over the period analyzed). This finding was true across all years of data analyzed, suggesting that the COVID-19 pandemic did not result in a quantifiable shift in care of skin cancer within the active-duty component to outside providers. One of the critical roles of dermatologists in the MHS is to diagnose and treat skin cancer, and our study suggested that the current global manning and staffing for MHS dermatologists may not be sufficient to meet the burden of skin cancers encountered within our active-duty troops, as only 61% are managed with DC. In particular, service members in more austere and/or overseas locations may not have ready access to a dermatologist.
The burden of skin cancer shifts dramatically when analyzing care of all other populations included in these data, including dependents of active-duty service members, retirees, and the category of “other” (ie, principally dependents of retirees). Within these populations, the rate of DC falls to 30%, with 70% of active-duty dependent care being deferred to network. The findings are even more noticeable for retirees and others within these 2 cohorts in all types of skin cancer analyzed, where DC only accounted for 5.2% of those skin cancers encountered and managed across TRICARE-eligible beneficiaries. For MM, BCC, and SCC, percentages of DC were 5.4%, 5.8%, and 3.5%, respectively. Although it is interesting to note the lower percentage of SCC managed via DC, our data did not allow for extrapolation as to why more SCC cases may be deferred to network. The shift to PC may align with DoD initiatives to increase the private sector’s involvement in military medicine and transition to civilianizing the MHS.14 In the end, the findings are remarkable, with approximately 95% of skin cancer care and management provided overall via PC.
These findings differ from previously published data regarding DC and PC from other specialty areas. Results from an analysis of DC vs PC for plastic surgery for the entire MHS from 2016 to 2019 found 83.2% of cases were deferred to network.15 A similar publication in the orthopedics literature examined TRICARE claims for patients who underwent total hip or knee arthroplasties between 2006 and 2019 and found 84.6% of cases were referred for PC. Notably, the authors utilized generalized linear models for cost analysis and found that DC was more expensive than PC, though this likely was a result of higher rates of hospital readmission within DC cases.16 Lastly, an article on the DC vs PC disposition of MHS patients with breast cancer from 2003 to 2008 found 46% of cases managed with DC vs 26.% with PC and 27.8% receiving a combination. In this case, the authors found a reduced cost associated with DC vs PC.17
Little additional literature exists regarding the costs of DC vs PC. An article published in 2016 designed to assess costs of DC vs PC showed that almost all military treatment facilities have higher costs than their private sector counterparts, with a few exceptions.18 This does not assess the costs of specific procedures, however, and only the overall cost to maintain a treatment facility. Importantly, this study was based on data from FY 2014 and has not been updated to reflect complex changes within the MHS system and the private health care system. Indeed, a US Government Accountability Office FY 2023 study highlighted staffing and efficiency issues within this transition to civilian medicine; subsequently, the 2024 President’s Budget suspended all planned clinical medical military end strength divestitures, underscoring the potential ineffectiveness of a civilianized MHS at meeting the health care needs of its beneficiaries.19,20 Future research on a national scale will be necessary to see if there is a reversal of this trend to PC and if doing so has any impact on access to DC for active-duty troops or active-duty dependents.
In addition to PC vs DC trends, we also can get a sense of the impact of the COVID pandemic restrictions on access to DC vs PC by assessing the change in rates seen in the data from the pre-COVID years (2017-2019) to the “post-COVID” years (2020-2022) included. Overall, rates of DC decreased uniformly from their already low percentages. In our study, rates of DC decreased from 5.8% in 2019 to 4.8% in 2022 for MM, from 6.6% to 4.3% for BCC, and from 4.2% to 2.9% for SCC. Although these changes seem small at first, they represent a 30.6% overall decrease in DC for BCC and an overall decrease of 55.4% in DC for SCC. Although our data do not allow us to extrapolate the real cost of this reduction across a nationwide health care system and more than 5 million care encounters, the financial and personal (ie, lost man-hours) costs of this decrease in DC likely are substantial.
In addition to costs, qualitative aspects that contribute to the burden of skin cancer include treatment-related morbidity, such as scarring, pain, and time spent away from family, work, and hobbies, as well as overall patient satisfaction with the quality of care they receive.21 Future work is critical to assess the real cost of this immense burden of PC for the treatment and management of skin cancers within the DoD beneficiary population.
Limitations—This study is limited by its observational nature. Given the mechanism of our data collection, we may have underestimated disease prevalence, as not all patients are seen for their diagnosis annually. Furthermore, reported demographic strata (eg, age, sex) were limited to those available and valid in the M2 reporting system. Finally, our study only collected data from those service members or former service members seen within the MHS and does not reflect any care rendered to those who are no longer active duty but did not officially retire from the military (ie, nonretired service members receiving care in the Veterans Affairs system for skin cancer).
Conclusion
We describe the annual burden of care for skin cancer in the MHS beneficiary population. Noteworthy findings observed were an overall decrease in beneficiaries being treated for skin cancer through DC; a decreasing annual prevalence of skin cancer diagnosis between 2019 and 2021, which may represent underdiagnosis or decreased follow-up in the setting of the COVID-19 pandemic; and a higher rate of skin cancer in the military beneficiary population compared to the civilian population.
- US Department of Defense. Military health. Accessed October 5, 2023. https://www.defense.gov/
- Wooten NR, Brittingham JA, Pitner RO, et al. Purchased behavioral health care received by Military Health System beneficiaries in civilian medical facilities, 2000-2014. Mil Med. 2018;183:E278-E290. doi:10.1093/milmed/usx101
- Riemenschneider K, Liu J, Powers JG. Skin cancer in the military: a systematic review of melanoma and nonmelanoma skin cancer incidence, prevention, and screening among active duty and veteran personnel. J Am Acad Dermatol. 2018;78:1185-1192. doi:10.1016/j.jaad.2017.11.062
- American Academy of Dermatology. Skin cancer. Updated April 22, 2022. Accessed April 17, 2024. https://www.aad.org/media/stats-skin-cancer
- Eide MJ, Krajenta R, Johnson D, et al. Identification of patients with nonmelanoma skin cancer using health maintenance organization claims data. Am J Epidemiol. 2010;171:123-128. doi:10.1093/aje/kwp352
- Kao SYZ, Ekwueme DU, Holman DM, et al. Economic burden of skin cancer treatment in the USA: an analysis of the Medical Expenditure Panel Survey Data, 2012-2018. Cancer Causes Control. 2023;34:205-212. doi:10.1007/s10552-022-01644-0
- Aggarwal P, Knabel P, Fleischer AB. United States burden of melanoma and non-melanoma skin cancer from 1990 to 2019. J Am Acad Dermatol. 2021;85:388-395. doi:10.1016/j.jaad.2021.03.109
- SEER*Explorer. SEER Incidence Data, November 2023 Submission (1975-2021). National Cancer Institute; 2024. Accessed April 17, 2024. https://seer.cancer.gov/statistics-network/explorer/application.html?site=53&data_type=1&graph_type=1&compareBy=sex&chk_sex_1=1&chk_sex_3=3&chk_sex_2=2&rate_type=2&race=1&age_range=1&advopt_precision=1&advopt_show_ci=on&hdn_view=1&advopt_show_apc=on&advopt_display=1
- Brown J, Kopf AW, Rigel DS, et al. Malignant melanoma in World War II veterans. Int J Dermatol. 1984;23:661-663. doi:10.1111/j.1365-4362.1984.tb01228.x
- Page WF, Whiteman D, Murphy M. A comparison of melanoma mortality among WWII veterans of the Pacific and European theaters. Ann Epidemiol. 2000;10:192-195. doi:10.1016/s1047-2797(99)00050-2
- Ramani ML, Bennett RG. High prevalence of skin cancer in World War II servicemen stationed in the Pacific theater. J Am Acad Dermatol. 1993;28:733-737. doi:10.1016/0190-9622(93)70102-Y
- Trepanowski N, Chang MS, Zhou G, et al. Delays in melanoma presentation during the COVID-19 pandemic: a nationwide multi-institutional cohort study. J Am Acad Dermatol. 2022;87:1217-1219. doi:10.1016/j.jaad.2022.06.031
- Gibbs A. COVID-19 shutdowns caused delays in melanoma diagnoses, study finds. OHSU News. August 4, 2022. Accessed April 17, 2024. https://news.ohsu.edu/2022/08/04/covid-19-shutdowns-caused-delays-in-melanoma-diagnoses-study-finds
- Kime P. Pentagon budget calls for ‘civilianizing’ military hospitals. Military Times. Published February 10, 2020. Accessed April 17, 2024. https://www.militarytimes.com/news/your-military/2020/02/10/pentagon-budget-calls-for-civilianizing-military-hospitals/
- O’Reilly EB, Norris E, Ortiz-Pomales YT, et al. A comparison of direct care at military medical treatment facilities with purchased care in plastic surgery operative volume. Plast Reconstr Surg Glob Open. 2022;10(10 suppl):124-125. doi:10.1097/01.GOX.0000898976.03344.62
- Haag A, Hosein S, Lyon S, et al. Outcomes for arthroplasties in military health: a retrospective analysis of direct versus purchased care. Mil Med. 2023;188(suppl 6):45-51. doi:10.1093/milmed/usac441
- Eaglehouse YL, Georg MW, Richard P, et al. Cost-efficiency of breast cancer care in the US Military Health System: an economic evaluation in direct and purchased care. Mil Med. 2019;184:e494-e501. doi:10.1093/milmed/usz025
- Lurie PM. Comparing the cost of military treatment facilities with private sector care. Institute for Defense Analyses; February 2016. Accessed April 17, 2024. https://www.ida.org/research-and-publications/publications/all/c/co/comparing-the-costs-of-military-treatment-facilities-with-private-sector-care
- Defense Health Program. Fiscal Year (FY) 2024 President’s Budget: Operation and Maintenance Procurement Research, Development, Test and Evaluation. Department of Defense; March 2023. Accessed April 17, 2024. https://comptroller.defense.gov/Portals/45/Documents/defbudget/fy2024/budget_justification/pdfs/09_Defense_Health_Program/00-DHP_Vols_I_II_and_III_PB24.pdf
- US Government Accountability Office. Defense Health Care. DOD should reevaluate market structure for military medical treatment facility management. Published August 21, 2023. Accessed April 17, 2024. https://www.gao.gov/products/gao-23-105441
- Rosenberg A, Cho S. We can do better at protecting our service members from skin cancer. Mil Med. 2022;187:311-313. doi:10.1093/milmed/usac198
This retrospective observational study investigates skin cancer prevalence and care patterns within the Military Health System (MHS) from 2017 to 2022. Utilizing the MHS Management Analysis and Reporting Tool (most commonly called M2), we analyzed more than 5 million patient encounters and documented skin cancer prevalence in the MHS beneficiary population utilizing available demographic data. Notable findings included an increased prevalence of skin cancer in the military population compared with the civilian population, a substantial decline in direct care (DC) visits at military treatment facilities compared with civilian purchased care (PC) visits, and a decreased total number of visits during COVID-19 restrictions.
The Military Health System (MHS) is a worldwide health care delivery system that serves 9.6 million beneficiaries, including military service members, retirees, and their families.1 Its mission is 2-fold: provide a medically ready force, and provide a medical benefit in keeping with the service and sacrifice of active-duty personnel, military retirees, and their families. For fiscal year (FY) 2022, active-duty service members and their families comprised 16.7% and 19.9% of beneficiaries, respectively, while retired service members and their families comprised 27% and 32% of beneficiaries, respectively.
The MHS operates under the authority of the Department of Defense (DoD) and is supported by an annual budget of approximately $50 billion.1 Health care provision within the MHS is managed by TRICARE regional networks.2 Within these networks, MHS beneficiaries may receive health care in 2 categories: direct care (DC) and purchased care (PC). Direct care is rendered in military treatment facilities by military or civilian providers contracted by the DoD, and PC is administered by civilian providers at civilian health care facilities within the TRICARE network, which is comprised of individual providers, clinics, and hospitals that have agreed to accept TRICARE beneficiaries.1 Purchased care is fee-for-service and paid for by the MHS. Of note, the MHS differs from the Veterans Affairs health care system in that the MHS through DC and PC sees only active-duty service members, active-duty dependents, retirees, and retirees’ dependents (primarily spouses), whereas Veterans Affairs sees only veterans (not necessarily retirees) discharged from military service with compensable medical conditions or disabilities.
Skin cancer presents a notable concern for the US Military, as the risk for skin cancer is thought to be higher than in the general population.3,4 This elevated risk is attributed to numerous factors inherent to active-duty service, including time spent in tropical environments, increased exposure to UV radiation, time spent at high altitudes, and decreased rates of sun-protective behaviors.3 Although numerous studies have explored the mechanisms that contribute to service members’ increased skin cancer risk, there are few (if any) that discuss the burden of skin cancer on the MHS and where its beneficiaries receive their skin cancer care. This study evaluated the burden of skin cancer within the MHS, as demonstrated by the period prevalence of skin cancer among its beneficiaries and the number and distribution of patient visits for skin cancer across both DC and PC from 2017 to 2022.
Methods
Data Collection—This retrospective observational study was designed to describe trends in outpatient visits with a skin cancer diagnosis and annual prevalence of skin cancer types in the MHS. Data are from all MHS beneficiaries who were eligible or enrolled in the analysis year. Our data source was the MHS Management Analysis and Reporting Tool (most commonly called M2), a query tool that contains the current and most recent 5 full FYs of Defense Health Agency corporate health care data including aggregated FY and calendar-year counts of MHS beneficiaries from 2017 to 2022 using encounter and claims data tables from both DC and PC. Data in M2 are coded using a pseudo-person identification number, and queries performed for this study were limited to de-identified visit and patient counts.
Skin cancer diagnoses were defined by relevant International Classification of Diseases, Tenth Revision, Clinical Modification (ICD-10-CM) codes recorded from outpatient visits in DC and PC. The M2 database was queried to find aggregate counts of visits and unique MHS beneficiaries with one or more diagnoses of a skin cancer type of interest (defined by relevant ICD-10-CM code) over the study period stratified by year and by patient demographic characteristics. Skin cancer types by ICD-10-CM code group included basal cell carcinoma (BCC), squamous cell carcinoma (SCC), malignant melanoma (MM), and other (including Merkel cell carcinoma and sebaceous carcinoma). Demographic strata included age, sex, military status (active duty, dependents of active duty, retired, or all others), sponsor military rank, and sponsor branch (army, air force, marine corps, or navy). Visit counts included diagnoses from any ICD position (for encounters that contained multiple ICD codes) to describe the total volume of care that addressed a diagnosed skin cancer. Counts of unique patients in prevalence analyses included relevant diagnoses in the primary ICD position only to increase the specificity of prevalence estimates.
Data Analysis—Descriptive analyses included the total number of outpatient visits with a skin cancer diagnosis in DC and PC over the study period, with percentages of total visits by year and by demographic strata. Separate analyses estimated annual prevalences of skin cancer types in the MHS by study year and within 2022 by demographic strata. Numerators in prevalence analyses were defined as the number of unique individuals with one or more relevant ICD codes in the analysis year. Denominators were defined as the total number of MHS beneficiaries in the analysis year and resulting period prevalences reported. Observed prevalences were qualitatively described, and trends were compared with prevalences in nonmilitary populations reported in the literature.
Ethics—This study was conducted as part of a study using secondary analyses of de-identified data from the M2 database. The study was reviewed and approved by the Walter Reed National Military Medical Center institutional review board.

Results
Encounter data were analyzed from a total of 5,374,348 visits between DC and PC over the study period for each cancer type of interest. Figures 1 and 2 show temporal trends in DC visits compared with PC visits in each beneficiary category. The percentage of total DC visits subsequently declined each year throughout the study period, with percentage decreases from 2017 to 2022 of 1.45% or 8200 fewer visits for MM, 3.41% or 7280 fewer visits for BCC, and 2.26% or 3673 fewer visits for SCC.

When stratified by beneficiary category, this trend remained consistent among dependents and retirees, with the most notable annual percentage decrease from 2019 to 2020. A higher proportion of younger adults and active-duty beneficiaries was seen in DC relative to PC, in which most visits were among retirees and others (primarily dependents of retirees, survivors, and Guard/Reserve on active duty, as well as inactive Guard/Reserve). No linear trends over time were apparent for active duty in DC and for dependents and retirees in PC. eTable 1 summarizes the demographic characteristics of MHS beneficiaries being seen in DC and PC over the study period for each cancer type of interest.

The Table shows the period prevalence of skin cancer diagnoses within the MHS beneficiary population from 2017 to 2022. These data were further analyzed by MM, BCC, and SCC (eTable 2) and demographics of interest for the year 2022. By beneficiary category, the period prevalence of MM was 0.08% in active duty, 0.06% in dependents, 0.48% in others, and 1.10% in retirees; the period prevalence of BCC was 0.12% in active duty, 0.07% in dependents, 0.91% in others, and 2.50% in retirees; and the period prevalence of SCC was 0.02% in active duty, 0.01% in dependents, 0.63% in others, and 1.87% in retirees. By sponsor branch, the period prevalence of MM was 0.35% in the army, 0.62% in the air force, 0.35% in the marine corps, and 0.65% in the navy; the period prevalence of BCC was 0.74% in the army, 1.30% in the air force, 0.74% in the marine corps, and 1.36% in the navy; and the period prevalence of SCC was 0.52% in the army, 0.92% in the air force, 0.51% in the marine corps, and 0.97% in the navy.


Comment
This study aimed to provide insight into the burden of skin cancer within the MHS beneficiary population and to identify temporal trends in where these beneficiaries receive their care. We examined patient encounter data from more than 9.6 million MHS beneficiaries.
The utilization of ICD codes from patient encounters to estimate the prevalence of nonmelanoma skin cancer (NMSC) has demonstrated a high positive predictive value. In one study, NMSC cases were confirmed in 96.5% of ICD code–identified patients.5 We presented an extensive collection of epidemiologic data on BCC and SCC, which posed unique challenges for tracking, as they are not reported to or monitored by cancer registries such as the Surveillance, Epidemiology, and End Results (SEER) Program.6
MHS Compared to the US Population—A study using the Global Burden of Disease 2019 database revealed an increasing trend in the incidence and prevalence of NMSC and melanoma since 1990. The same study found the period prevalence in 2019 of MM, SCC, and BCC in the general US population to be 0.13%, 0.31%, and 0.05%, respectively.7 In contrast, among MHS beneficiaries, we observed a higher prevalence in the same year, with figures of 0.66% for MM, 0.72% for SCC, and 1.02% for BCC. According to the SEER database, the period prevalence of MM within the general US population in 2020 was 0.4%.8 That same year, we identified a higher period prevalence of MM—0.54%—within the MHS beneficiary population. Specifically, within the MHS retiree population, the prevalence in 2022 was double that of the general MHS population, with a rate of 1.10%, underscoring the importance of skin cancer screening in older, at-risk adult populations. Prior studies similarly found increased rates of skin cancer within the military beneficiary population. Further studies are needed to compare age-adjusted rates in the MHS vs US population.9-11
COVID-19 Trends—Our data showed an overall decreasing prevalence of skin cancer in the MHS from 2019 to 2021. We suspect that the apparent decrease in skin cancer prevalence may be attributed to underdiagnosis from COVID-19 pandemic restrictions. During that time, many dermatology clinics at military treatment facilities underwent temporary closures, and some dermatologists were sent on nondermatologic utilization tours. Likewise, a US multi-institutional study described declining rates of new melanomas from 2020 to 2021, with an increased proportion of patient presentations with advanced melanoma, suggesting an underdiagnosis of melanoma cases during pandemic restrictions. That study also noted an increased rate of patient-identified melanomas and a decreased rate of provider-identified melanomas during that time.12 Contributing factors may include excess hospital demand, increased patient complexity and acute care needs, and long outpatient clinic backlogs during this time.13Financial Burden—Over our 5-year study period, there were 5,374,348 patient encounters addressing skin cancer, both in DC and PC (Figures 1 and 2; eTable 1). In 2016 to 2018, the average annual cost of treating skin cancer in the US civilian, noninstitutionalized population was $1243 for NMSC (BCC and SCC) and $2430 for melanoma.6 Using this metric, the estimated total cost of care rendered in the MHS in 2018 for NMSC and melanoma was $202,510,803 and $156,516,300, respectively.
Trends in DC vs PC—In the years examined, we found a notable decrease in the number of beneficiaries receiving treatment for MM, BCC, and SCC in DC. Simultaneously, there has been an increase in the number of beneficiaries receiving PC for BCC and SCC, though this trend was not apparent for MM.
Our data provided interesting insights into the percentage of PC compared with DC offered within the MHS. Importantly, our findings suggested that the majority of skin cancer in active-duty service members is managed with DC within the military treatment facility setting (61% DC management over the period analyzed). This finding was true across all years of data analyzed, suggesting that the COVID-19 pandemic did not result in a quantifiable shift in care of skin cancer within the active-duty component to outside providers. One of the critical roles of dermatologists in the MHS is to diagnose and treat skin cancer, and our study suggested that the current global manning and staffing for MHS dermatologists may not be sufficient to meet the burden of skin cancers encountered within our active-duty troops, as only 61% are managed with DC. In particular, service members in more austere and/or overseas locations may not have ready access to a dermatologist.
The burden of skin cancer shifts dramatically when analyzing care of all other populations included in these data, including dependents of active-duty service members, retirees, and the category of “other” (ie, principally dependents of retirees). Within these populations, the rate of DC falls to 30%, with 70% of active-duty dependent care being deferred to network. The findings are even more noticeable for retirees and others within these 2 cohorts in all types of skin cancer analyzed, where DC only accounted for 5.2% of those skin cancers encountered and managed across TRICARE-eligible beneficiaries. For MM, BCC, and SCC, percentages of DC were 5.4%, 5.8%, and 3.5%, respectively. Although it is interesting to note the lower percentage of SCC managed via DC, our data did not allow for extrapolation as to why more SCC cases may be deferred to network. The shift to PC may align with DoD initiatives to increase the private sector’s involvement in military medicine and transition to civilianizing the MHS.14 In the end, the findings are remarkable, with approximately 95% of skin cancer care and management provided overall via PC.
These findings differ from previously published data regarding DC and PC from other specialty areas. Results from an analysis of DC vs PC for plastic surgery for the entire MHS from 2016 to 2019 found 83.2% of cases were deferred to network.15 A similar publication in the orthopedics literature examined TRICARE claims for patients who underwent total hip or knee arthroplasties between 2006 and 2019 and found 84.6% of cases were referred for PC. Notably, the authors utilized generalized linear models for cost analysis and found that DC was more expensive than PC, though this likely was a result of higher rates of hospital readmission within DC cases.16 Lastly, an article on the DC vs PC disposition of MHS patients with breast cancer from 2003 to 2008 found 46% of cases managed with DC vs 26.% with PC and 27.8% receiving a combination. In this case, the authors found a reduced cost associated with DC vs PC.17
Little additional literature exists regarding the costs of DC vs PC. An article published in 2016 designed to assess costs of DC vs PC showed that almost all military treatment facilities have higher costs than their private sector counterparts, with a few exceptions.18 This does not assess the costs of specific procedures, however, and only the overall cost to maintain a treatment facility. Importantly, this study was based on data from FY 2014 and has not been updated to reflect complex changes within the MHS system and the private health care system. Indeed, a US Government Accountability Office FY 2023 study highlighted staffing and efficiency issues within this transition to civilian medicine; subsequently, the 2024 President’s Budget suspended all planned clinical medical military end strength divestitures, underscoring the potential ineffectiveness of a civilianized MHS at meeting the health care needs of its beneficiaries.19,20 Future research on a national scale will be necessary to see if there is a reversal of this trend to PC and if doing so has any impact on access to DC for active-duty troops or active-duty dependents.
In addition to PC vs DC trends, we also can get a sense of the impact of the COVID pandemic restrictions on access to DC vs PC by assessing the change in rates seen in the data from the pre-COVID years (2017-2019) to the “post-COVID” years (2020-2022) included. Overall, rates of DC decreased uniformly from their already low percentages. In our study, rates of DC decreased from 5.8% in 2019 to 4.8% in 2022 for MM, from 6.6% to 4.3% for BCC, and from 4.2% to 2.9% for SCC. Although these changes seem small at first, they represent a 30.6% overall decrease in DC for BCC and an overall decrease of 55.4% in DC for SCC. Although our data do not allow us to extrapolate the real cost of this reduction across a nationwide health care system and more than 5 million care encounters, the financial and personal (ie, lost man-hours) costs of this decrease in DC likely are substantial.
In addition to costs, qualitative aspects that contribute to the burden of skin cancer include treatment-related morbidity, such as scarring, pain, and time spent away from family, work, and hobbies, as well as overall patient satisfaction with the quality of care they receive.21 Future work is critical to assess the real cost of this immense burden of PC for the treatment and management of skin cancers within the DoD beneficiary population.
Limitations—This study is limited by its observational nature. Given the mechanism of our data collection, we may have underestimated disease prevalence, as not all patients are seen for their diagnosis annually. Furthermore, reported demographic strata (eg, age, sex) were limited to those available and valid in the M2 reporting system. Finally, our study only collected data from those service members or former service members seen within the MHS and does not reflect any care rendered to those who are no longer active duty but did not officially retire from the military (ie, nonretired service members receiving care in the Veterans Affairs system for skin cancer).
Conclusion
We describe the annual burden of care for skin cancer in the MHS beneficiary population. Noteworthy findings observed were an overall decrease in beneficiaries being treated for skin cancer through DC; a decreasing annual prevalence of skin cancer diagnosis between 2019 and 2021, which may represent underdiagnosis or decreased follow-up in the setting of the COVID-19 pandemic; and a higher rate of skin cancer in the military beneficiary population compared to the civilian population.
This retrospective observational study investigates skin cancer prevalence and care patterns within the Military Health System (MHS) from 2017 to 2022. Utilizing the MHS Management Analysis and Reporting Tool (most commonly called M2), we analyzed more than 5 million patient encounters and documented skin cancer prevalence in the MHS beneficiary population utilizing available demographic data. Notable findings included an increased prevalence of skin cancer in the military population compared with the civilian population, a substantial decline in direct care (DC) visits at military treatment facilities compared with civilian purchased care (PC) visits, and a decreased total number of visits during COVID-19 restrictions.
The Military Health System (MHS) is a worldwide health care delivery system that serves 9.6 million beneficiaries, including military service members, retirees, and their families.1 Its mission is 2-fold: provide a medically ready force, and provide a medical benefit in keeping with the service and sacrifice of active-duty personnel, military retirees, and their families. For fiscal year (FY) 2022, active-duty service members and their families comprised 16.7% and 19.9% of beneficiaries, respectively, while retired service members and their families comprised 27% and 32% of beneficiaries, respectively.
The MHS operates under the authority of the Department of Defense (DoD) and is supported by an annual budget of approximately $50 billion.1 Health care provision within the MHS is managed by TRICARE regional networks.2 Within these networks, MHS beneficiaries may receive health care in 2 categories: direct care (DC) and purchased care (PC). Direct care is rendered in military treatment facilities by military or civilian providers contracted by the DoD, and PC is administered by civilian providers at civilian health care facilities within the TRICARE network, which is comprised of individual providers, clinics, and hospitals that have agreed to accept TRICARE beneficiaries.1 Purchased care is fee-for-service and paid for by the MHS. Of note, the MHS differs from the Veterans Affairs health care system in that the MHS through DC and PC sees only active-duty service members, active-duty dependents, retirees, and retirees’ dependents (primarily spouses), whereas Veterans Affairs sees only veterans (not necessarily retirees) discharged from military service with compensable medical conditions or disabilities.
Skin cancer presents a notable concern for the US Military, as the risk for skin cancer is thought to be higher than in the general population.3,4 This elevated risk is attributed to numerous factors inherent to active-duty service, including time spent in tropical environments, increased exposure to UV radiation, time spent at high altitudes, and decreased rates of sun-protective behaviors.3 Although numerous studies have explored the mechanisms that contribute to service members’ increased skin cancer risk, there are few (if any) that discuss the burden of skin cancer on the MHS and where its beneficiaries receive their skin cancer care. This study evaluated the burden of skin cancer within the MHS, as demonstrated by the period prevalence of skin cancer among its beneficiaries and the number and distribution of patient visits for skin cancer across both DC and PC from 2017 to 2022.
Methods
Data Collection—This retrospective observational study was designed to describe trends in outpatient visits with a skin cancer diagnosis and annual prevalence of skin cancer types in the MHS. Data are from all MHS beneficiaries who were eligible or enrolled in the analysis year. Our data source was the MHS Management Analysis and Reporting Tool (most commonly called M2), a query tool that contains the current and most recent 5 full FYs of Defense Health Agency corporate health care data including aggregated FY and calendar-year counts of MHS beneficiaries from 2017 to 2022 using encounter and claims data tables from both DC and PC. Data in M2 are coded using a pseudo-person identification number, and queries performed for this study were limited to de-identified visit and patient counts.
Skin cancer diagnoses were defined by relevant International Classification of Diseases, Tenth Revision, Clinical Modification (ICD-10-CM) codes recorded from outpatient visits in DC and PC. The M2 database was queried to find aggregate counts of visits and unique MHS beneficiaries with one or more diagnoses of a skin cancer type of interest (defined by relevant ICD-10-CM code) over the study period stratified by year and by patient demographic characteristics. Skin cancer types by ICD-10-CM code group included basal cell carcinoma (BCC), squamous cell carcinoma (SCC), malignant melanoma (MM), and other (including Merkel cell carcinoma and sebaceous carcinoma). Demographic strata included age, sex, military status (active duty, dependents of active duty, retired, or all others), sponsor military rank, and sponsor branch (army, air force, marine corps, or navy). Visit counts included diagnoses from any ICD position (for encounters that contained multiple ICD codes) to describe the total volume of care that addressed a diagnosed skin cancer. Counts of unique patients in prevalence analyses included relevant diagnoses in the primary ICD position only to increase the specificity of prevalence estimates.
Data Analysis—Descriptive analyses included the total number of outpatient visits with a skin cancer diagnosis in DC and PC over the study period, with percentages of total visits by year and by demographic strata. Separate analyses estimated annual prevalences of skin cancer types in the MHS by study year and within 2022 by demographic strata. Numerators in prevalence analyses were defined as the number of unique individuals with one or more relevant ICD codes in the analysis year. Denominators were defined as the total number of MHS beneficiaries in the analysis year and resulting period prevalences reported. Observed prevalences were qualitatively described, and trends were compared with prevalences in nonmilitary populations reported in the literature.
Ethics—This study was conducted as part of a study using secondary analyses of de-identified data from the M2 database. The study was reviewed and approved by the Walter Reed National Military Medical Center institutional review board.

Results
Encounter data were analyzed from a total of 5,374,348 visits between DC and PC over the study period for each cancer type of interest. Figures 1 and 2 show temporal trends in DC visits compared with PC visits in each beneficiary category. The percentage of total DC visits subsequently declined each year throughout the study period, with percentage decreases from 2017 to 2022 of 1.45% or 8200 fewer visits for MM, 3.41% or 7280 fewer visits for BCC, and 2.26% or 3673 fewer visits for SCC.

When stratified by beneficiary category, this trend remained consistent among dependents and retirees, with the most notable annual percentage decrease from 2019 to 2020. A higher proportion of younger adults and active-duty beneficiaries was seen in DC relative to PC, in which most visits were among retirees and others (primarily dependents of retirees, survivors, and Guard/Reserve on active duty, as well as inactive Guard/Reserve). No linear trends over time were apparent for active duty in DC and for dependents and retirees in PC. eTable 1 summarizes the demographic characteristics of MHS beneficiaries being seen in DC and PC over the study period for each cancer type of interest.

The Table shows the period prevalence of skin cancer diagnoses within the MHS beneficiary population from 2017 to 2022. These data were further analyzed by MM, BCC, and SCC (eTable 2) and demographics of interest for the year 2022. By beneficiary category, the period prevalence of MM was 0.08% in active duty, 0.06% in dependents, 0.48% in others, and 1.10% in retirees; the period prevalence of BCC was 0.12% in active duty, 0.07% in dependents, 0.91% in others, and 2.50% in retirees; and the period prevalence of SCC was 0.02% in active duty, 0.01% in dependents, 0.63% in others, and 1.87% in retirees. By sponsor branch, the period prevalence of MM was 0.35% in the army, 0.62% in the air force, 0.35% in the marine corps, and 0.65% in the navy; the period prevalence of BCC was 0.74% in the army, 1.30% in the air force, 0.74% in the marine corps, and 1.36% in the navy; and the period prevalence of SCC was 0.52% in the army, 0.92% in the air force, 0.51% in the marine corps, and 0.97% in the navy.


Comment
This study aimed to provide insight into the burden of skin cancer within the MHS beneficiary population and to identify temporal trends in where these beneficiaries receive their care. We examined patient encounter data from more than 9.6 million MHS beneficiaries.
The utilization of ICD codes from patient encounters to estimate the prevalence of nonmelanoma skin cancer (NMSC) has demonstrated a high positive predictive value. In one study, NMSC cases were confirmed in 96.5% of ICD code–identified patients.5 We presented an extensive collection of epidemiologic data on BCC and SCC, which posed unique challenges for tracking, as they are not reported to or monitored by cancer registries such as the Surveillance, Epidemiology, and End Results (SEER) Program.6
MHS Compared to the US Population—A study using the Global Burden of Disease 2019 database revealed an increasing trend in the incidence and prevalence of NMSC and melanoma since 1990. The same study found the period prevalence in 2019 of MM, SCC, and BCC in the general US population to be 0.13%, 0.31%, and 0.05%, respectively.7 In contrast, among MHS beneficiaries, we observed a higher prevalence in the same year, with figures of 0.66% for MM, 0.72% for SCC, and 1.02% for BCC. According to the SEER database, the period prevalence of MM within the general US population in 2020 was 0.4%.8 That same year, we identified a higher period prevalence of MM—0.54%—within the MHS beneficiary population. Specifically, within the MHS retiree population, the prevalence in 2022 was double that of the general MHS population, with a rate of 1.10%, underscoring the importance of skin cancer screening in older, at-risk adult populations. Prior studies similarly found increased rates of skin cancer within the military beneficiary population. Further studies are needed to compare age-adjusted rates in the MHS vs US population.9-11
COVID-19 Trends—Our data showed an overall decreasing prevalence of skin cancer in the MHS from 2019 to 2021. We suspect that the apparent decrease in skin cancer prevalence may be attributed to underdiagnosis from COVID-19 pandemic restrictions. During that time, many dermatology clinics at military treatment facilities underwent temporary closures, and some dermatologists were sent on nondermatologic utilization tours. Likewise, a US multi-institutional study described declining rates of new melanomas from 2020 to 2021, with an increased proportion of patient presentations with advanced melanoma, suggesting an underdiagnosis of melanoma cases during pandemic restrictions. That study also noted an increased rate of patient-identified melanomas and a decreased rate of provider-identified melanomas during that time.12 Contributing factors may include excess hospital demand, increased patient complexity and acute care needs, and long outpatient clinic backlogs during this time.13Financial Burden—Over our 5-year study period, there were 5,374,348 patient encounters addressing skin cancer, both in DC and PC (Figures 1 and 2; eTable 1). In 2016 to 2018, the average annual cost of treating skin cancer in the US civilian, noninstitutionalized population was $1243 for NMSC (BCC and SCC) and $2430 for melanoma.6 Using this metric, the estimated total cost of care rendered in the MHS in 2018 for NMSC and melanoma was $202,510,803 and $156,516,300, respectively.
Trends in DC vs PC—In the years examined, we found a notable decrease in the number of beneficiaries receiving treatment for MM, BCC, and SCC in DC. Simultaneously, there has been an increase in the number of beneficiaries receiving PC for BCC and SCC, though this trend was not apparent for MM.
Our data provided interesting insights into the percentage of PC compared with DC offered within the MHS. Importantly, our findings suggested that the majority of skin cancer in active-duty service members is managed with DC within the military treatment facility setting (61% DC management over the period analyzed). This finding was true across all years of data analyzed, suggesting that the COVID-19 pandemic did not result in a quantifiable shift in care of skin cancer within the active-duty component to outside providers. One of the critical roles of dermatologists in the MHS is to diagnose and treat skin cancer, and our study suggested that the current global manning and staffing for MHS dermatologists may not be sufficient to meet the burden of skin cancers encountered within our active-duty troops, as only 61% are managed with DC. In particular, service members in more austere and/or overseas locations may not have ready access to a dermatologist.
The burden of skin cancer shifts dramatically when analyzing care of all other populations included in these data, including dependents of active-duty service members, retirees, and the category of “other” (ie, principally dependents of retirees). Within these populations, the rate of DC falls to 30%, with 70% of active-duty dependent care being deferred to network. The findings are even more noticeable for retirees and others within these 2 cohorts in all types of skin cancer analyzed, where DC only accounted for 5.2% of those skin cancers encountered and managed across TRICARE-eligible beneficiaries. For MM, BCC, and SCC, percentages of DC were 5.4%, 5.8%, and 3.5%, respectively. Although it is interesting to note the lower percentage of SCC managed via DC, our data did not allow for extrapolation as to why more SCC cases may be deferred to network. The shift to PC may align with DoD initiatives to increase the private sector’s involvement in military medicine and transition to civilianizing the MHS.14 In the end, the findings are remarkable, with approximately 95% of skin cancer care and management provided overall via PC.
These findings differ from previously published data regarding DC and PC from other specialty areas. Results from an analysis of DC vs PC for plastic surgery for the entire MHS from 2016 to 2019 found 83.2% of cases were deferred to network.15 A similar publication in the orthopedics literature examined TRICARE claims for patients who underwent total hip or knee arthroplasties between 2006 and 2019 and found 84.6% of cases were referred for PC. Notably, the authors utilized generalized linear models for cost analysis and found that DC was more expensive than PC, though this likely was a result of higher rates of hospital readmission within DC cases.16 Lastly, an article on the DC vs PC disposition of MHS patients with breast cancer from 2003 to 2008 found 46% of cases managed with DC vs 26.% with PC and 27.8% receiving a combination. In this case, the authors found a reduced cost associated with DC vs PC.17
Little additional literature exists regarding the costs of DC vs PC. An article published in 2016 designed to assess costs of DC vs PC showed that almost all military treatment facilities have higher costs than their private sector counterparts, with a few exceptions.18 This does not assess the costs of specific procedures, however, and only the overall cost to maintain a treatment facility. Importantly, this study was based on data from FY 2014 and has not been updated to reflect complex changes within the MHS system and the private health care system. Indeed, a US Government Accountability Office FY 2023 study highlighted staffing and efficiency issues within this transition to civilian medicine; subsequently, the 2024 President’s Budget suspended all planned clinical medical military end strength divestitures, underscoring the potential ineffectiveness of a civilianized MHS at meeting the health care needs of its beneficiaries.19,20 Future research on a national scale will be necessary to see if there is a reversal of this trend to PC and if doing so has any impact on access to DC for active-duty troops or active-duty dependents.
In addition to PC vs DC trends, we also can get a sense of the impact of the COVID pandemic restrictions on access to DC vs PC by assessing the change in rates seen in the data from the pre-COVID years (2017-2019) to the “post-COVID” years (2020-2022) included. Overall, rates of DC decreased uniformly from their already low percentages. In our study, rates of DC decreased from 5.8% in 2019 to 4.8% in 2022 for MM, from 6.6% to 4.3% for BCC, and from 4.2% to 2.9% for SCC. Although these changes seem small at first, they represent a 30.6% overall decrease in DC for BCC and an overall decrease of 55.4% in DC for SCC. Although our data do not allow us to extrapolate the real cost of this reduction across a nationwide health care system and more than 5 million care encounters, the financial and personal (ie, lost man-hours) costs of this decrease in DC likely are substantial.
In addition to costs, qualitative aspects that contribute to the burden of skin cancer include treatment-related morbidity, such as scarring, pain, and time spent away from family, work, and hobbies, as well as overall patient satisfaction with the quality of care they receive.21 Future work is critical to assess the real cost of this immense burden of PC for the treatment and management of skin cancers within the DoD beneficiary population.
Limitations—This study is limited by its observational nature. Given the mechanism of our data collection, we may have underestimated disease prevalence, as not all patients are seen for their diagnosis annually. Furthermore, reported demographic strata (eg, age, sex) were limited to those available and valid in the M2 reporting system. Finally, our study only collected data from those service members or former service members seen within the MHS and does not reflect any care rendered to those who are no longer active duty but did not officially retire from the military (ie, nonretired service members receiving care in the Veterans Affairs system for skin cancer).
Conclusion
We describe the annual burden of care for skin cancer in the MHS beneficiary population. Noteworthy findings observed were an overall decrease in beneficiaries being treated for skin cancer through DC; a decreasing annual prevalence of skin cancer diagnosis between 2019 and 2021, which may represent underdiagnosis or decreased follow-up in the setting of the COVID-19 pandemic; and a higher rate of skin cancer in the military beneficiary population compared to the civilian population.
- US Department of Defense. Military health. Accessed October 5, 2023. https://www.defense.gov/
- Wooten NR, Brittingham JA, Pitner RO, et al. Purchased behavioral health care received by Military Health System beneficiaries in civilian medical facilities, 2000-2014. Mil Med. 2018;183:E278-E290. doi:10.1093/milmed/usx101
- Riemenschneider K, Liu J, Powers JG. Skin cancer in the military: a systematic review of melanoma and nonmelanoma skin cancer incidence, prevention, and screening among active duty and veteran personnel. J Am Acad Dermatol. 2018;78:1185-1192. doi:10.1016/j.jaad.2017.11.062
- American Academy of Dermatology. Skin cancer. Updated April 22, 2022. Accessed April 17, 2024. https://www.aad.org/media/stats-skin-cancer
- Eide MJ, Krajenta R, Johnson D, et al. Identification of patients with nonmelanoma skin cancer using health maintenance organization claims data. Am J Epidemiol. 2010;171:123-128. doi:10.1093/aje/kwp352
- Kao SYZ, Ekwueme DU, Holman DM, et al. Economic burden of skin cancer treatment in the USA: an analysis of the Medical Expenditure Panel Survey Data, 2012-2018. Cancer Causes Control. 2023;34:205-212. doi:10.1007/s10552-022-01644-0
- Aggarwal P, Knabel P, Fleischer AB. United States burden of melanoma and non-melanoma skin cancer from 1990 to 2019. J Am Acad Dermatol. 2021;85:388-395. doi:10.1016/j.jaad.2021.03.109
- SEER*Explorer. SEER Incidence Data, November 2023 Submission (1975-2021). National Cancer Institute; 2024. Accessed April 17, 2024. https://seer.cancer.gov/statistics-network/explorer/application.html?site=53&data_type=1&graph_type=1&compareBy=sex&chk_sex_1=1&chk_sex_3=3&chk_sex_2=2&rate_type=2&race=1&age_range=1&advopt_precision=1&advopt_show_ci=on&hdn_view=1&advopt_show_apc=on&advopt_display=1
- Brown J, Kopf AW, Rigel DS, et al. Malignant melanoma in World War II veterans. Int J Dermatol. 1984;23:661-663. doi:10.1111/j.1365-4362.1984.tb01228.x
- Page WF, Whiteman D, Murphy M. A comparison of melanoma mortality among WWII veterans of the Pacific and European theaters. Ann Epidemiol. 2000;10:192-195. doi:10.1016/s1047-2797(99)00050-2
- Ramani ML, Bennett RG. High prevalence of skin cancer in World War II servicemen stationed in the Pacific theater. J Am Acad Dermatol. 1993;28:733-737. doi:10.1016/0190-9622(93)70102-Y
- Trepanowski N, Chang MS, Zhou G, et al. Delays in melanoma presentation during the COVID-19 pandemic: a nationwide multi-institutional cohort study. J Am Acad Dermatol. 2022;87:1217-1219. doi:10.1016/j.jaad.2022.06.031
- Gibbs A. COVID-19 shutdowns caused delays in melanoma diagnoses, study finds. OHSU News. August 4, 2022. Accessed April 17, 2024. https://news.ohsu.edu/2022/08/04/covid-19-shutdowns-caused-delays-in-melanoma-diagnoses-study-finds
- Kime P. Pentagon budget calls for ‘civilianizing’ military hospitals. Military Times. Published February 10, 2020. Accessed April 17, 2024. https://www.militarytimes.com/news/your-military/2020/02/10/pentagon-budget-calls-for-civilianizing-military-hospitals/
- O’Reilly EB, Norris E, Ortiz-Pomales YT, et al. A comparison of direct care at military medical treatment facilities with purchased care in plastic surgery operative volume. Plast Reconstr Surg Glob Open. 2022;10(10 suppl):124-125. doi:10.1097/01.GOX.0000898976.03344.62
- Haag A, Hosein S, Lyon S, et al. Outcomes for arthroplasties in military health: a retrospective analysis of direct versus purchased care. Mil Med. 2023;188(suppl 6):45-51. doi:10.1093/milmed/usac441
- Eaglehouse YL, Georg MW, Richard P, et al. Cost-efficiency of breast cancer care in the US Military Health System: an economic evaluation in direct and purchased care. Mil Med. 2019;184:e494-e501. doi:10.1093/milmed/usz025
- Lurie PM. Comparing the cost of military treatment facilities with private sector care. Institute for Defense Analyses; February 2016. Accessed April 17, 2024. https://www.ida.org/research-and-publications/publications/all/c/co/comparing-the-costs-of-military-treatment-facilities-with-private-sector-care
- Defense Health Program. Fiscal Year (FY) 2024 President’s Budget: Operation and Maintenance Procurement Research, Development, Test and Evaluation. Department of Defense; March 2023. Accessed April 17, 2024. https://comptroller.defense.gov/Portals/45/Documents/defbudget/fy2024/budget_justification/pdfs/09_Defense_Health_Program/00-DHP_Vols_I_II_and_III_PB24.pdf
- US Government Accountability Office. Defense Health Care. DOD should reevaluate market structure for military medical treatment facility management. Published August 21, 2023. Accessed April 17, 2024. https://www.gao.gov/products/gao-23-105441
- Rosenberg A, Cho S. We can do better at protecting our service members from skin cancer. Mil Med. 2022;187:311-313. doi:10.1093/milmed/usac198
- US Department of Defense. Military health. Accessed October 5, 2023. https://www.defense.gov/
- Wooten NR, Brittingham JA, Pitner RO, et al. Purchased behavioral health care received by Military Health System beneficiaries in civilian medical facilities, 2000-2014. Mil Med. 2018;183:E278-E290. doi:10.1093/milmed/usx101
- Riemenschneider K, Liu J, Powers JG. Skin cancer in the military: a systematic review of melanoma and nonmelanoma skin cancer incidence, prevention, and screening among active duty and veteran personnel. J Am Acad Dermatol. 2018;78:1185-1192. doi:10.1016/j.jaad.2017.11.062
- American Academy of Dermatology. Skin cancer. Updated April 22, 2022. Accessed April 17, 2024. https://www.aad.org/media/stats-skin-cancer
- Eide MJ, Krajenta R, Johnson D, et al. Identification of patients with nonmelanoma skin cancer using health maintenance organization claims data. Am J Epidemiol. 2010;171:123-128. doi:10.1093/aje/kwp352
- Kao SYZ, Ekwueme DU, Holman DM, et al. Economic burden of skin cancer treatment in the USA: an analysis of the Medical Expenditure Panel Survey Data, 2012-2018. Cancer Causes Control. 2023;34:205-212. doi:10.1007/s10552-022-01644-0
- Aggarwal P, Knabel P, Fleischer AB. United States burden of melanoma and non-melanoma skin cancer from 1990 to 2019. J Am Acad Dermatol. 2021;85:388-395. doi:10.1016/j.jaad.2021.03.109
- SEER*Explorer. SEER Incidence Data, November 2023 Submission (1975-2021). National Cancer Institute; 2024. Accessed April 17, 2024. https://seer.cancer.gov/statistics-network/explorer/application.html?site=53&data_type=1&graph_type=1&compareBy=sex&chk_sex_1=1&chk_sex_3=3&chk_sex_2=2&rate_type=2&race=1&age_range=1&advopt_precision=1&advopt_show_ci=on&hdn_view=1&advopt_show_apc=on&advopt_display=1
- Brown J, Kopf AW, Rigel DS, et al. Malignant melanoma in World War II veterans. Int J Dermatol. 1984;23:661-663. doi:10.1111/j.1365-4362.1984.tb01228.x
- Page WF, Whiteman D, Murphy M. A comparison of melanoma mortality among WWII veterans of the Pacific and European theaters. Ann Epidemiol. 2000;10:192-195. doi:10.1016/s1047-2797(99)00050-2
- Ramani ML, Bennett RG. High prevalence of skin cancer in World War II servicemen stationed in the Pacific theater. J Am Acad Dermatol. 1993;28:733-737. doi:10.1016/0190-9622(93)70102-Y
- Trepanowski N, Chang MS, Zhou G, et al. Delays in melanoma presentation during the COVID-19 pandemic: a nationwide multi-institutional cohort study. J Am Acad Dermatol. 2022;87:1217-1219. doi:10.1016/j.jaad.2022.06.031
- Gibbs A. COVID-19 shutdowns caused delays in melanoma diagnoses, study finds. OHSU News. August 4, 2022. Accessed April 17, 2024. https://news.ohsu.edu/2022/08/04/covid-19-shutdowns-caused-delays-in-melanoma-diagnoses-study-finds
- Kime P. Pentagon budget calls for ‘civilianizing’ military hospitals. Military Times. Published February 10, 2020. Accessed April 17, 2024. https://www.militarytimes.com/news/your-military/2020/02/10/pentagon-budget-calls-for-civilianizing-military-hospitals/
- O’Reilly EB, Norris E, Ortiz-Pomales YT, et al. A comparison of direct care at military medical treatment facilities with purchased care in plastic surgery operative volume. Plast Reconstr Surg Glob Open. 2022;10(10 suppl):124-125. doi:10.1097/01.GOX.0000898976.03344.62
- Haag A, Hosein S, Lyon S, et al. Outcomes for arthroplasties in military health: a retrospective analysis of direct versus purchased care. Mil Med. 2023;188(suppl 6):45-51. doi:10.1093/milmed/usac441
- Eaglehouse YL, Georg MW, Richard P, et al. Cost-efficiency of breast cancer care in the US Military Health System: an economic evaluation in direct and purchased care. Mil Med. 2019;184:e494-e501. doi:10.1093/milmed/usz025
- Lurie PM. Comparing the cost of military treatment facilities with private sector care. Institute for Defense Analyses; February 2016. Accessed April 17, 2024. https://www.ida.org/research-and-publications/publications/all/c/co/comparing-the-costs-of-military-treatment-facilities-with-private-sector-care
- Defense Health Program. Fiscal Year (FY) 2024 President’s Budget: Operation and Maintenance Procurement Research, Development, Test and Evaluation. Department of Defense; March 2023. Accessed April 17, 2024. https://comptroller.defense.gov/Portals/45/Documents/defbudget/fy2024/budget_justification/pdfs/09_Defense_Health_Program/00-DHP_Vols_I_II_and_III_PB24.pdf
- US Government Accountability Office. Defense Health Care. DOD should reevaluate market structure for military medical treatment facility management. Published August 21, 2023. Accessed April 17, 2024. https://www.gao.gov/products/gao-23-105441
- Rosenberg A, Cho S. We can do better at protecting our service members from skin cancer. Mil Med. 2022;187:311-313. doi:10.1093/milmed/usac198
PRACTICE POINTS
- Study data showed an overall decreasing prevalence of skin cancer in the Military Health System (MHS) from 2019 to 2021, possibly attributable to underdiagnosis resulting from the COVID-19 pandemic. Providers should be mindful of this trend when screening patients who have experienced interruptions in care.
- An overall increased prevalence of skin cancer was noted in the military beneficiary population compared with publicly available civilian data—and thus this diagnosis should be given special consideration within this population.
The Clinical Utility of Teledermatology in Triaging and Diagnosing Skin Malignancies: Case Series
With the increasing utilization of telemedicine since the COVID-19 pandemic, it is critical that clinicians have an appropriate understanding of the application of virtual care resources, including teledermatology. We present a case series of 3 patients to demonstrate the clinical utility of teledermatology in reducing the time to diagnosis of various rare and/or aggressive cutaneous malignancies, including Merkel cell carcinoma, malignant melanoma, and atypical fibroxanthoma. Cases were obtained from one large Midwestern medical center during the month of July 2021. Each case presented includes a description of the initial teledermatology presentation and reviews the clinical timeline from initial consultation submission to in-person clinic visit with lesion biopsy. This case series demonstrates real-world examples of how teledermatology can be utilized to expedite the care of specific vulnerable patient populations.
Teledermatology is a rapidly growing digital resource with specific utility in triaging patients to determine those requiring in-person evaluation for early and accurate detection of skin malignancies. Approximately one-third of teledermatology consultations result in face-to-face clinical encounters, with malignant neoplasms being the leading cause for biopsy.1,2 For specific populations, such as geriatric and immunocompromised patients, teledermatology may serve as a valuable tool, particularly in the wake of the COVID-19 pandemic. Furthermore, telemedicine may aid in addressing health disparities within the field of medicine and ultimately may improve access to care for vulnerable populations.3 Along with increasing access to specific subspecialty expertise, the use of teledermatology may reduce health care costs and improve the overall quality of care delivered to patients.4,5
We describe the clinical utility of teledermatology in triaging and diagnosing skin malignancies through a series of 3 cases obtained from digital image review at one large Midwestern medical center during the month of July 2021. Three unique cases with a final diagnosis of a rare or aggressive skin cancer were selected as examples, including a 75-year-old man with Merkle cell carcinoma, a 55-year-old man with aggressive pT3b malignant melanoma, and a 72-year-old man with an atypical fibroxanthoma. A clinical timeline of each case is presented, including the time intervals from initial image submission to image review, image submission to face-to-face clinical encounter, and image submission to final diagnosis. In all cases, the primary care provider submitted an order for teledermatology, and the teledermatology team obtained the images.
Case Series
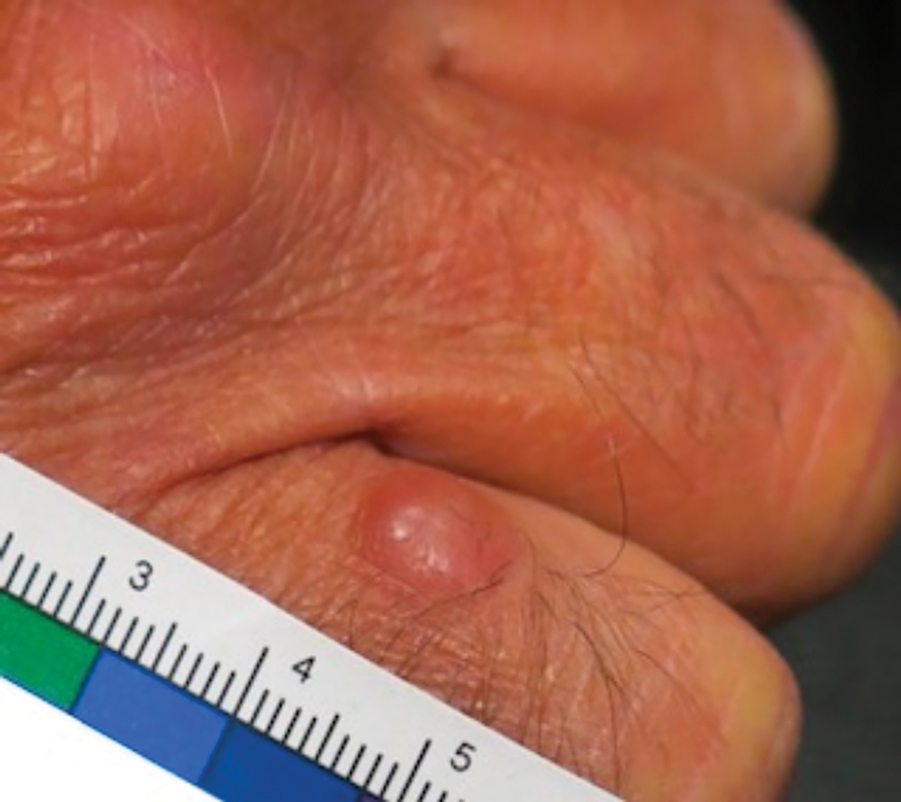
Patient 1—Images of the right hand of a 75-year-old man with a medical history of basal cell carcinoma were submitted for teledermatology consultation utilizing store-and-forward image-capturing technology (day 1). The patient history provided with image submission indicated that the lesion had been present for 6 months and there were no associated symptoms. Clinical imaging demonstrated a pink-red pearly papule located on the proximal fourth digit of the dorsal aspect of the right hand (Figure 1). One day following the teledermatology request (day 2), the patient’s case was reviewed and triaged for an in-person visit. The patient was brought to clinic on day 34, and a biopsy was performed. On day 36, dermatopathology results indicated a diagnosis of Merkel cell carcinoma. On day 37, the patient was referred to surgical oncology, and on day 44, the patient underwent an initial surgical oncology visit with a plan for wide local excision of the right fourth digit with right axillary sentinel lymph node biopsy.
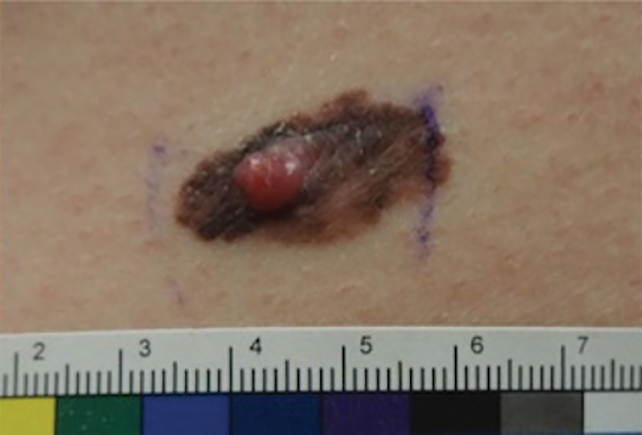
Patient 2—Images of the left flank of a 55-year-old man were submitted for teledermatology consultation via store-and-forward technology (day 1). A patient history provided with the image indicated that the lesion had been present for months to years and there were no associated symptoms, but the lesion recently had changed in color and size. Teledermatology images were reviewed on day 3 and demonstrated a 2- to 3-cm brown plaque on the left flank with color variegation and a prominent red papule protruding centrally (Figure 2). The patient was scheduled for an urgent in-person visit with biopsy. On day 6, the patient presented to clinic and an excision biopsy was performed. Dermatopathology was ordered with a RUSH indication, with results on day 7 revealing a pT3b malignant melanoma. An urgent consultation to surgical oncology was placed on the same day, and the patient underwent an initial surgical oncology visit on day 24 with a plan for wide local excision with left axillary and inguinal sentinel lymph node biopsy.
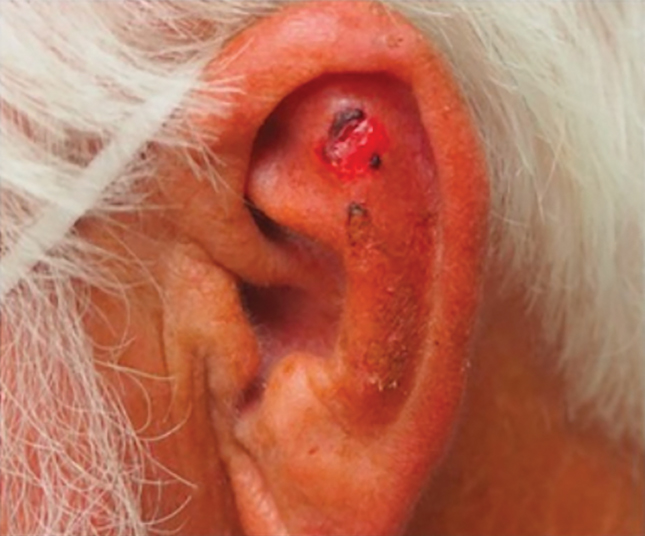
Patient 3—Images of the left ear of a 72-year-old man were submitted for teledermatology consultation utilizing review via store-and-forward technology (day 1). A patient history indicated that the lesion had been present for 3 months with associated bleeding. Image review demonstrated a solitary pearly pink papule located on the crura of the antihelix (Figure 3). Initial teledermatology consultation was reviewed on day 2 with notification of the need for in-person evaluation. The patient presented to clinic on day 33 for a biopsy, with dermatopathology results on day 36 consistent with an atypical fibroxanthoma. The patient was scheduled for Mohs micrographic surgery on day 37 and underwent surgical treatment on day 64.
Comment
Teledermatology consultations from all patients demonstrated adequate image quality to be able to evaluate the lesion of concern and yielded a request for in-person evaluation with possible biopsy (Table). In this case series, the average time interval from teledermatology consultation placement to teledermatology image report was 2 days (range, 1–3 days). The average time from teledermatology consultation placement to face-to-face encounter with biopsy was 24.3 days for the 3 cases presented in this series (range, 6–34 days). The initial surgical oncology visits took place an average of 34 days after the initial teledermatology consultation was placed for the 2 patients requiring referral (44 days for patient 1; 24 days for patient 2). For patient 3, Mohs micrographic surgery was required for treatment, which was scheduled by day 37 and subsequently performed on day 64.
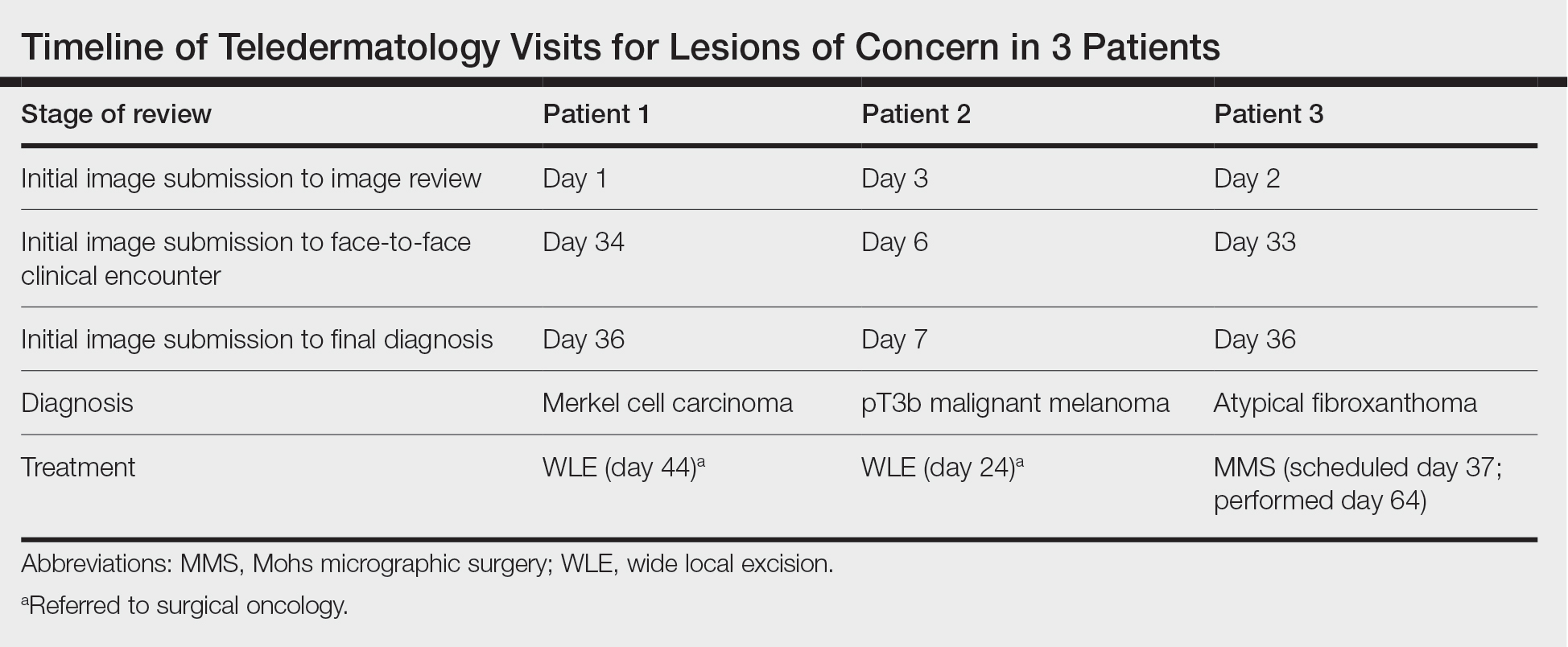
When specifically looking at the diagnosis of cutaneous malignancies, studies have found that the incidence of skin cancer detection is similar for teledermatology compared to in-person clinic visits.6,7 Creighton-Smith et al6 performed a retrospective cohort study comparing prebiopsy and postbiopsy diagnostic accuracy and detection rates of skin cancer between store-and-forward technology and face-to-face consultation. When adjusting for possible compounding factors including personal and family history of skin cancer, there was no notable difference in detection rates of any skin cancer, including melanoma and nonmelanoma skin cancers. Furthermore, the 2 cohorts of patients were found to have similar prebiopsy and postbiopsy diagnostic concordance, with similar times from consultation being placed to requested biopsy and time from biopsy to final treatment.6
Clarke et al7 similarly analyzed the accuracy of store-and-forward teledermatology and found that there was overall concordance in diagnosis when comparing clinical dermatologists to teledermatologists. Moreover, when melanocytic lesions were excluded from the study, the decision to biopsy did not differ substantially.7
Areas of further study include determining what percentage of teledermatology lesions of concern for malignancy were proven to be skin cancer after in-person evaluation and biopsy, as well as investigating the effectiveness of teledermatology for melanocytic lesions, which frequently are removed from analysis in large-scale teledermatology studies.
Although teledermatology has substantial clinical utility and may serve as a great resource for specific populations, including geriatric patients and those who are immunocompromised, it is important to recognize notable limitations. Specifically, brief history and image review should not serve as replacements for a face-to-face visit with physical examination in cases where the diagnosis remains uncertain or when high-risk skin malignancies are suspected or included in the differential. Certain aggressive cutaneous malignancies such as Merkel cell carcinoma may appear as less aggressive via teledermatology due to restrictions of technology.
Conclusion
Teledermatology has had a major impact on the way health care is delivered to patients and may increase access to care, reducing unnecessary in-person visits and decreasing the number of in-person visit no-shows. With the appropriate use of a brief clinical history and image review, teledermatology can be effective to evaluate specific lesions of concern. We report 3 unique cases identified during a 1-month period at a large Midwestern medical center. These cases serve as important examples of the application of teledermatology in reducing the time to diagnosis of aggressive skin malignancies. Further research on the clinical utility of teledermatology is warranted.
Acknowledgments—The authors thank the additional providers from the University of Wisconsin and William S. Middleton Memorial Veterans Hospital (both in Madison, Wisconsin) involved in the medical care of the patients included in this case series.
- Bianchi MG, Santos A, Cordioli E. Benefits of teledermatology for geriatric patients: population-based cross-sectional study. J Med Internet Res. 2020;22:E16700.
- Mortimer S, Rosin A. A retrospective review of incidental malignancies in veterans seen for face-to-face follow-up after teledermatology consultation. J Am Acad Dermatol. 2021;84:1130-1132.
- Costello CM, Cumsky HJL, Maly CJ, et al. Improving access to care through the establishment of a local, teledermatology network. Telemed J E Health. 2020;26:935-940. doi:10.1089/tmj.2019.0051
- Lee JJ, English JC 3rd. Teledermatology: a review and update. Am J Clin Dermatol. 2018;19:253-260. doi:10.1007/s40257-017-0317-6
- Hadeler E, Beer J, Nouri K. The influence of teledermatology on health care access and equity. J Am Acad Dermatol. 2021;84:E219-E220. doi:10.1016/j.jaad.2020.12.036
- Creighton-Smith M, Murgia RD 3rd, Konnikov N, et al. Incidence of melanoma and keratinocytic carcinomas in patients evaluated by store-and-forward teledermatology vs dermatology clinic. Int J Dermatol. 2017;56:1026-1031. doi:10.1111/ijd.13672
- Clarke EL, Reichenberg JS, Ahmed AM, et al. The utility of teledermatology in the evaluation of skin lesions. J Telemed Telecare. 2023;29:382-389. doi:10.1177/1357633X20987423
With the increasing utilization of telemedicine since the COVID-19 pandemic, it is critical that clinicians have an appropriate understanding of the application of virtual care resources, including teledermatology. We present a case series of 3 patients to demonstrate the clinical utility of teledermatology in reducing the time to diagnosis of various rare and/or aggressive cutaneous malignancies, including Merkel cell carcinoma, malignant melanoma, and atypical fibroxanthoma. Cases were obtained from one large Midwestern medical center during the month of July 2021. Each case presented includes a description of the initial teledermatology presentation and reviews the clinical timeline from initial consultation submission to in-person clinic visit with lesion biopsy. This case series demonstrates real-world examples of how teledermatology can be utilized to expedite the care of specific vulnerable patient populations.
Teledermatology is a rapidly growing digital resource with specific utility in triaging patients to determine those requiring in-person evaluation for early and accurate detection of skin malignancies. Approximately one-third of teledermatology consultations result in face-to-face clinical encounters, with malignant neoplasms being the leading cause for biopsy.1,2 For specific populations, such as geriatric and immunocompromised patients, teledermatology may serve as a valuable tool, particularly in the wake of the COVID-19 pandemic. Furthermore, telemedicine may aid in addressing health disparities within the field of medicine and ultimately may improve access to care for vulnerable populations.3 Along with increasing access to specific subspecialty expertise, the use of teledermatology may reduce health care costs and improve the overall quality of care delivered to patients.4,5
We describe the clinical utility of teledermatology in triaging and diagnosing skin malignancies through a series of 3 cases obtained from digital image review at one large Midwestern medical center during the month of July 2021. Three unique cases with a final diagnosis of a rare or aggressive skin cancer were selected as examples, including a 75-year-old man with Merkle cell carcinoma, a 55-year-old man with aggressive pT3b malignant melanoma, and a 72-year-old man with an atypical fibroxanthoma. A clinical timeline of each case is presented, including the time intervals from initial image submission to image review, image submission to face-to-face clinical encounter, and image submission to final diagnosis. In all cases, the primary care provider submitted an order for teledermatology, and the teledermatology team obtained the images.
Case Series
Patient 1—Images of the right hand of a 75-year-old man with a medical history of basal cell carcinoma were submitted for teledermatology consultation utilizing store-and-forward image-capturing technology (day 1). The patient history provided with image submission indicated that the lesion had been present for 6 months and there were no associated symptoms. Clinical imaging demonstrated a pink-red pearly papule located on the proximal fourth digit of the dorsal aspect of the right hand (Figure 1). One day following the teledermatology request (day 2), the patient’s case was reviewed and triaged for an in-person visit. The patient was brought to clinic on day 34, and a biopsy was performed. On day 36, dermatopathology results indicated a diagnosis of Merkel cell carcinoma. On day 37, the patient was referred to surgical oncology, and on day 44, the patient underwent an initial surgical oncology visit with a plan for wide local excision of the right fourth digit with right axillary sentinel lymph node biopsy.

Patient 2—Images of the left flank of a 55-year-old man were submitted for teledermatology consultation via store-and-forward technology (day 1). A patient history provided with the image indicated that the lesion had been present for months to years and there were no associated symptoms, but the lesion recently had changed in color and size. Teledermatology images were reviewed on day 3 and demonstrated a 2- to 3-cm brown plaque on the left flank with color variegation and a prominent red papule protruding centrally (Figure 2). The patient was scheduled for an urgent in-person visit with biopsy. On day 6, the patient presented to clinic and an excision biopsy was performed. Dermatopathology was ordered with a RUSH indication, with results on day 7 revealing a pT3b malignant melanoma. An urgent consultation to surgical oncology was placed on the same day, and the patient underwent an initial surgical oncology visit on day 24 with a plan for wide local excision with left axillary and inguinal sentinel lymph node biopsy.

Patient 3—Images of the left ear of a 72-year-old man were submitted for teledermatology consultation utilizing review via store-and-forward technology (day 1). A patient history indicated that the lesion had been present for 3 months with associated bleeding. Image review demonstrated a solitary pearly pink papule located on the crura of the antihelix (Figure 3). Initial teledermatology consultation was reviewed on day 2 with notification of the need for in-person evaluation. The patient presented to clinic on day 33 for a biopsy, with dermatopathology results on day 36 consistent with an atypical fibroxanthoma. The patient was scheduled for Mohs micrographic surgery on day 37 and underwent surgical treatment on day 64.

Comment
Teledermatology consultations from all patients demonstrated adequate image quality to be able to evaluate the lesion of concern and yielded a request for in-person evaluation with possible biopsy (Table). In this case series, the average time interval from teledermatology consultation placement to teledermatology image report was 2 days (range, 1–3 days). The average time from teledermatology consultation placement to face-to-face encounter with biopsy was 24.3 days for the 3 cases presented in this series (range, 6–34 days). The initial surgical oncology visits took place an average of 34 days after the initial teledermatology consultation was placed for the 2 patients requiring referral (44 days for patient 1; 24 days for patient 2). For patient 3, Mohs micrographic surgery was required for treatment, which was scheduled by day 37 and subsequently performed on day 64.

When specifically looking at the diagnosis of cutaneous malignancies, studies have found that the incidence of skin cancer detection is similar for teledermatology compared to in-person clinic visits.6,7 Creighton-Smith et al6 performed a retrospective cohort study comparing prebiopsy and postbiopsy diagnostic accuracy and detection rates of skin cancer between store-and-forward technology and face-to-face consultation. When adjusting for possible compounding factors including personal and family history of skin cancer, there was no notable difference in detection rates of any skin cancer, including melanoma and nonmelanoma skin cancers. Furthermore, the 2 cohorts of patients were found to have similar prebiopsy and postbiopsy diagnostic concordance, with similar times from consultation being placed to requested biopsy and time from biopsy to final treatment.6
Clarke et al7 similarly analyzed the accuracy of store-and-forward teledermatology and found that there was overall concordance in diagnosis when comparing clinical dermatologists to teledermatologists. Moreover, when melanocytic lesions were excluded from the study, the decision to biopsy did not differ substantially.7
Areas of further study include determining what percentage of teledermatology lesions of concern for malignancy were proven to be skin cancer after in-person evaluation and biopsy, as well as investigating the effectiveness of teledermatology for melanocytic lesions, which frequently are removed from analysis in large-scale teledermatology studies.
Although teledermatology has substantial clinical utility and may serve as a great resource for specific populations, including geriatric patients and those who are immunocompromised, it is important to recognize notable limitations. Specifically, brief history and image review should not serve as replacements for a face-to-face visit with physical examination in cases where the diagnosis remains uncertain or when high-risk skin malignancies are suspected or included in the differential. Certain aggressive cutaneous malignancies such as Merkel cell carcinoma may appear as less aggressive via teledermatology due to restrictions of technology.
Conclusion
Teledermatology has had a major impact on the way health care is delivered to patients and may increase access to care, reducing unnecessary in-person visits and decreasing the number of in-person visit no-shows. With the appropriate use of a brief clinical history and image review, teledermatology can be effective to evaluate specific lesions of concern. We report 3 unique cases identified during a 1-month period at a large Midwestern medical center. These cases serve as important examples of the application of teledermatology in reducing the time to diagnosis of aggressive skin malignancies. Further research on the clinical utility of teledermatology is warranted.
Acknowledgments—The authors thank the additional providers from the University of Wisconsin and William S. Middleton Memorial Veterans Hospital (both in Madison, Wisconsin) involved in the medical care of the patients included in this case series.
With the increasing utilization of telemedicine since the COVID-19 pandemic, it is critical that clinicians have an appropriate understanding of the application of virtual care resources, including teledermatology. We present a case series of 3 patients to demonstrate the clinical utility of teledermatology in reducing the time to diagnosis of various rare and/or aggressive cutaneous malignancies, including Merkel cell carcinoma, malignant melanoma, and atypical fibroxanthoma. Cases were obtained from one large Midwestern medical center during the month of July 2021. Each case presented includes a description of the initial teledermatology presentation and reviews the clinical timeline from initial consultation submission to in-person clinic visit with lesion biopsy. This case series demonstrates real-world examples of how teledermatology can be utilized to expedite the care of specific vulnerable patient populations.
Teledermatology is a rapidly growing digital resource with specific utility in triaging patients to determine those requiring in-person evaluation for early and accurate detection of skin malignancies. Approximately one-third of teledermatology consultations result in face-to-face clinical encounters, with malignant neoplasms being the leading cause for biopsy.1,2 For specific populations, such as geriatric and immunocompromised patients, teledermatology may serve as a valuable tool, particularly in the wake of the COVID-19 pandemic. Furthermore, telemedicine may aid in addressing health disparities within the field of medicine and ultimately may improve access to care for vulnerable populations.3 Along with increasing access to specific subspecialty expertise, the use of teledermatology may reduce health care costs and improve the overall quality of care delivered to patients.4,5
We describe the clinical utility of teledermatology in triaging and diagnosing skin malignancies through a series of 3 cases obtained from digital image review at one large Midwestern medical center during the month of July 2021. Three unique cases with a final diagnosis of a rare or aggressive skin cancer were selected as examples, including a 75-year-old man with Merkle cell carcinoma, a 55-year-old man with aggressive pT3b malignant melanoma, and a 72-year-old man with an atypical fibroxanthoma. A clinical timeline of each case is presented, including the time intervals from initial image submission to image review, image submission to face-to-face clinical encounter, and image submission to final diagnosis. In all cases, the primary care provider submitted an order for teledermatology, and the teledermatology team obtained the images.
Case Series
Patient 1—Images of the right hand of a 75-year-old man with a medical history of basal cell carcinoma were submitted for teledermatology consultation utilizing store-and-forward image-capturing technology (day 1). The patient history provided with image submission indicated that the lesion had been present for 6 months and there were no associated symptoms. Clinical imaging demonstrated a pink-red pearly papule located on the proximal fourth digit of the dorsal aspect of the right hand (Figure 1). One day following the teledermatology request (day 2), the patient’s case was reviewed and triaged for an in-person visit. The patient was brought to clinic on day 34, and a biopsy was performed. On day 36, dermatopathology results indicated a diagnosis of Merkel cell carcinoma. On day 37, the patient was referred to surgical oncology, and on day 44, the patient underwent an initial surgical oncology visit with a plan for wide local excision of the right fourth digit with right axillary sentinel lymph node biopsy.

Patient 2—Images of the left flank of a 55-year-old man were submitted for teledermatology consultation via store-and-forward technology (day 1). A patient history provided with the image indicated that the lesion had been present for months to years and there were no associated symptoms, but the lesion recently had changed in color and size. Teledermatology images were reviewed on day 3 and demonstrated a 2- to 3-cm brown plaque on the left flank with color variegation and a prominent red papule protruding centrally (Figure 2). The patient was scheduled for an urgent in-person visit with biopsy. On day 6, the patient presented to clinic and an excision biopsy was performed. Dermatopathology was ordered with a RUSH indication, with results on day 7 revealing a pT3b malignant melanoma. An urgent consultation to surgical oncology was placed on the same day, and the patient underwent an initial surgical oncology visit on day 24 with a plan for wide local excision with left axillary and inguinal sentinel lymph node biopsy.

Patient 3—Images of the left ear of a 72-year-old man were submitted for teledermatology consultation utilizing review via store-and-forward technology (day 1). A patient history indicated that the lesion had been present for 3 months with associated bleeding. Image review demonstrated a solitary pearly pink papule located on the crura of the antihelix (Figure 3). Initial teledermatology consultation was reviewed on day 2 with notification of the need for in-person evaluation. The patient presented to clinic on day 33 for a biopsy, with dermatopathology results on day 36 consistent with an atypical fibroxanthoma. The patient was scheduled for Mohs micrographic surgery on day 37 and underwent surgical treatment on day 64.

Comment
Teledermatology consultations from all patients demonstrated adequate image quality to be able to evaluate the lesion of concern and yielded a request for in-person evaluation with possible biopsy (Table). In this case series, the average time interval from teledermatology consultation placement to teledermatology image report was 2 days (range, 1–3 days). The average time from teledermatology consultation placement to face-to-face encounter with biopsy was 24.3 days for the 3 cases presented in this series (range, 6–34 days). The initial surgical oncology visits took place an average of 34 days after the initial teledermatology consultation was placed for the 2 patients requiring referral (44 days for patient 1; 24 days for patient 2). For patient 3, Mohs micrographic surgery was required for treatment, which was scheduled by day 37 and subsequently performed on day 64.

When specifically looking at the diagnosis of cutaneous malignancies, studies have found that the incidence of skin cancer detection is similar for teledermatology compared to in-person clinic visits.6,7 Creighton-Smith et al6 performed a retrospective cohort study comparing prebiopsy and postbiopsy diagnostic accuracy and detection rates of skin cancer between store-and-forward technology and face-to-face consultation. When adjusting for possible compounding factors including personal and family history of skin cancer, there was no notable difference in detection rates of any skin cancer, including melanoma and nonmelanoma skin cancers. Furthermore, the 2 cohorts of patients were found to have similar prebiopsy and postbiopsy diagnostic concordance, with similar times from consultation being placed to requested biopsy and time from biopsy to final treatment.6
Clarke et al7 similarly analyzed the accuracy of store-and-forward teledermatology and found that there was overall concordance in diagnosis when comparing clinical dermatologists to teledermatologists. Moreover, when melanocytic lesions were excluded from the study, the decision to biopsy did not differ substantially.7
Areas of further study include determining what percentage of teledermatology lesions of concern for malignancy were proven to be skin cancer after in-person evaluation and biopsy, as well as investigating the effectiveness of teledermatology for melanocytic lesions, which frequently are removed from analysis in large-scale teledermatology studies.
Although teledermatology has substantial clinical utility and may serve as a great resource for specific populations, including geriatric patients and those who are immunocompromised, it is important to recognize notable limitations. Specifically, brief history and image review should not serve as replacements for a face-to-face visit with physical examination in cases where the diagnosis remains uncertain or when high-risk skin malignancies are suspected or included in the differential. Certain aggressive cutaneous malignancies such as Merkel cell carcinoma may appear as less aggressive via teledermatology due to restrictions of technology.
Conclusion
Teledermatology has had a major impact on the way health care is delivered to patients and may increase access to care, reducing unnecessary in-person visits and decreasing the number of in-person visit no-shows. With the appropriate use of a brief clinical history and image review, teledermatology can be effective to evaluate specific lesions of concern. We report 3 unique cases identified during a 1-month period at a large Midwestern medical center. These cases serve as important examples of the application of teledermatology in reducing the time to diagnosis of aggressive skin malignancies. Further research on the clinical utility of teledermatology is warranted.
Acknowledgments—The authors thank the additional providers from the University of Wisconsin and William S. Middleton Memorial Veterans Hospital (both in Madison, Wisconsin) involved in the medical care of the patients included in this case series.
- Bianchi MG, Santos A, Cordioli E. Benefits of teledermatology for geriatric patients: population-based cross-sectional study. J Med Internet Res. 2020;22:E16700.
- Mortimer S, Rosin A. A retrospective review of incidental malignancies in veterans seen for face-to-face follow-up after teledermatology consultation. J Am Acad Dermatol. 2021;84:1130-1132.
- Costello CM, Cumsky HJL, Maly CJ, et al. Improving access to care through the establishment of a local, teledermatology network. Telemed J E Health. 2020;26:935-940. doi:10.1089/tmj.2019.0051
- Lee JJ, English JC 3rd. Teledermatology: a review and update. Am J Clin Dermatol. 2018;19:253-260. doi:10.1007/s40257-017-0317-6
- Hadeler E, Beer J, Nouri K. The influence of teledermatology on health care access and equity. J Am Acad Dermatol. 2021;84:E219-E220. doi:10.1016/j.jaad.2020.12.036
- Creighton-Smith M, Murgia RD 3rd, Konnikov N, et al. Incidence of melanoma and keratinocytic carcinomas in patients evaluated by store-and-forward teledermatology vs dermatology clinic. Int J Dermatol. 2017;56:1026-1031. doi:10.1111/ijd.13672
- Clarke EL, Reichenberg JS, Ahmed AM, et al. The utility of teledermatology in the evaluation of skin lesions. J Telemed Telecare. 2023;29:382-389. doi:10.1177/1357633X20987423
- Bianchi MG, Santos A, Cordioli E. Benefits of teledermatology for geriatric patients: population-based cross-sectional study. J Med Internet Res. 2020;22:E16700.
- Mortimer S, Rosin A. A retrospective review of incidental malignancies in veterans seen for face-to-face follow-up after teledermatology consultation. J Am Acad Dermatol. 2021;84:1130-1132.
- Costello CM, Cumsky HJL, Maly CJ, et al. Improving access to care through the establishment of a local, teledermatology network. Telemed J E Health. 2020;26:935-940. doi:10.1089/tmj.2019.0051
- Lee JJ, English JC 3rd. Teledermatology: a review and update. Am J Clin Dermatol. 2018;19:253-260. doi:10.1007/s40257-017-0317-6
- Hadeler E, Beer J, Nouri K. The influence of teledermatology on health care access and equity. J Am Acad Dermatol. 2021;84:E219-E220. doi:10.1016/j.jaad.2020.12.036
- Creighton-Smith M, Murgia RD 3rd, Konnikov N, et al. Incidence of melanoma and keratinocytic carcinomas in patients evaluated by store-and-forward teledermatology vs dermatology clinic. Int J Dermatol. 2017;56:1026-1031. doi:10.1111/ijd.13672
- Clarke EL, Reichenberg JS, Ahmed AM, et al. The utility of teledermatology in the evaluation of skin lesions. J Telemed Telecare. 2023;29:382-389. doi:10.1177/1357633X20987423
Practice Points
- Teledermatology via store-and-forward technology has been demonstrated to be effective in assessing and triaging various cutaneous malignancies.
- The use of teledermatology has increased because of the COVID-19 pandemic and may be useful for specific vulnerable populations.
- When used appropriately, teledermatology may function as a useful resource to triage patients requiring in-person evaluation for the diagnosis of aggressive skin malignancies and may aid in reducing the time to diagnosis of various skin cancers.
The Long, Controversial Search for a ‘Cancer Microbiome’
Last year, the controversy heightened when experts questioned a high-profile study — a 2020 analysis claiming that the tumors of 33 different cancers had their own unique microbiomes — on whether the “signature” of these bacterial compositions could help diagnose cancer.
The incident renewed the spotlight on “tumor microbiomes” because of the bold claims of the original paper and the strongly worded refutations of those claims. The broader field has focused primarily on ways the body’s microbiome interacts with cancers and cancer treatment.
This controversy has highlighted the challenges of making headway in a field where researchers may not even have the tools yet to puzzle-out the wide-ranging implications the microbiome holds for cancer diagnosis and treatment.
But it is also part of a provocative question within that larger field: whether tumors in the body, far from the natural microbiome in the gut, have their own thriving communities of bacteria, viruses, and fungi. And, if they do, how do those tumor microbiomes affect the development and progression of the cancer and the effectiveness of cancer therapies?
Cancer Controversy
The evidence is undeniable that some microbes can directly cause certain cancers and that the human gut microbiome can influence the effectiveness of certain therapies. Beyond that established science, however, the research has raised as many questions as answers about what we do and don’t know about microbiota and cancer.
The only confirmed microbiomes are on the skin and in the gut, mouth, and vagina, which are all areas with an easy direct route for bacteria to enter and grow in or on the body. A series of papers in recent years have suggested that other internal organs, and tumors within them, may have their own microbiomes.
“Whether microbes exist in tumors of internal organs beyond body surfaces exposed to the environment is a different matter,” said Ivan Vujkovic-Cvijin, PhD, an assistant professor of biomedical sciences and gastroenterology at Cedars-Sinai Medical Center in Los Angeles, whose lab studies how human gut microbes affect inflammatory diseases. “We’ve only recently had the tools to study that question on a molecular level, and the reported results have been conflicting.”
For example, research allegedly identified microbiota in the human placenta nearly one decade ago. But subsequent research contradicted those claims and showed that the source of the “placental microbiome” was actually contamination. Subsequent similar studies for other parts of the body faced the same scrutiny and, often, eventual debunking.
“Most likely, our immune system has undergone selective pressure to eliminate everything that crosses the gut barrier because there’s not much benefit to the body to have bacteria run amok in our internal organs,” Dr. Vujkovic-Cvijin said. “That can only disrupt the functioning of our tissues, to have an external organism living inside them.”
The controversy that erupted last summer, surrounding research from the lab of Rob Knight, PhD, at the University of California, San Diego, centered on a slightly different but related question: Could tumors harbor their own microbiomes?
This news organization spoke with two of the authors who published a paper contesting Dr. Knight’s findings: Steven Salzberg, PhD, a professor of biomedical engineering at John Hopkins Medicine, Baltimore, Maryland, and Abraham Gihawi, PhD, a research fellow at Norwich Medical School at the University of East Anglia in the United Kingdom.
Dr. Salzberg described two major problems with Dr. Knight’s study.
“What they found were false positives because of contamination in the database and flaws in their methods,” Dr. Salzberg said. “I can’t prove there’s no cancer microbiome, but I can say the cancer microbiomes that they reported don’t exist because the species they were finding aren’t there.”
Dr. Knight disagrees with Dr. Salzberg’s findings, noting that Dr. Salzberg and his co-authors did not examine the publicly available databases used in his study. In a written response, he said that his team’s examination of the database revealed that less than 1% of the microbial genomes overlapped with human ones and that removing them did not change their findings.
Dr. Knight also noted that his team could still “distinguish cancer types by their microbiome” even after running their analysis without the technique that Dr. Salzberg found fault with.
Dr. Salzberg said that the database linked above is not the one Dr. Knight’s study used, however. “The primary database in their study was never made public (it’s too large, they said), and it has/had about 69,000 genomes,” Dr. Salzberg said by email. “But even if we did, this is irrelevant. He’s trying to distract from the primary errors in their study,” which Dr. Salzberg said Dr. Knight’s team has not addressed.
The critiques Dr. Salzberg raised have been leveled at other studies investigating microbiomes specifically within tumors and independent of the body’s microbiome.
For example, a 2019 study in Nature described a fungal microbiome in pancreatic cancer that a Nature paper 4 years later directly contradicted, citing flaws that invalidated the original findings. A different 2019 study in Cell examined pancreatic tumor microbiota and patient outcomes, but it’s unclear whether the microorganisms moved from the gut to the pancreas or “constitute a durably colonized community that lives inside the tumor,” which remains a matter of debate, Dr. Vujkovic-Cvijin said.
A 2020 study in Science suggested diverse microbial communities in seven tumor types, but those findings were similarly called into question. That study stated that “bacteria were first detected in human tumors more than 100 years ago” and that “bacteria are well-known residents in human tumors,” but Dr. Salzberg considers those statements misleading.
It’s true that bacteria and viruses have been detected in tumors because “there’s very good evidence that an acute infection caused by a very small number of viruses and bacteria can cause a tumor,” Dr. Salzberg said. Human papillomavirus, for example, can cause six different types of cancer. Inflammation and ulcers caused by Helicobacter pylori may progress to stomach cancer, and Fusobacterium nucleatum and Enterococcus faecalis have been shown to contribute to colorectal cancer. Those examples differ from a microbiome; this “a community of bacteria and possibly other microscopic bugs, like fungi, that are happily living in the tumor” the same way microbes reside in our guts, he said.
Dr. Knight said that many bacteria his team identified “have been confirmed independently in subsequent work.” He acknowledged, however, that more research is needed.
Several of the contested studies above were among a lengthy list that Dr. Knight provided, noting that most of the disagreements “have two sides to them, and critiques from one particular group does not immediately invalidate a reported finding.”
Yet, many of the papers Dr. Knight listed are precisely the types that skeptics like Dr. Salzberg believe are too flawed to draw reliable conclusions.
“I think many agree that microbes may exist within tumors that are exposed to the environment, like tumors of the skin, gut, and mouth,” Dr. Vujkovic-Cvijin said. It’s less clear, however, whether tumors further from the body’s microbiome harbor any microbes or where they came from if they do. Microbial signals in organs elsewhere in the body become faint quickly, he said.
Underdeveloped Technology
Though Dr. Salzberg said that the concept of a tumor microbiome is “implausible” because there’s no easy route for bacteria to reach internal organs, it’s unclear whether scientists have the technology yet to adequately answer this question.
For one thing, samples in these types of studies are typically “ultra-low biomass samples, where the signal — the amount of microbes in the sample — is so low that it’s comparable to how much would be expected to be found in reagents and environmental contamination through processing,” Dr. Vujkovic-Cvijin explained. Many polymerases used to amplify a DNA signal, for example, are made in bacteria and may retain trace amounts identified in these studies.
Dr. Knight agreed that low biomass is a challenge in this field but is not an unsurmountable one.
Another challenge is that study samples, as with Dr. Knight’s work, were collected during routine surgeries without the intent to find a microbial signal. Simply using a scalpel to cut through the skin means cutting through a layer of bacteria, and surgery rooms are not designed to eliminate all bacteria. Some work has even shown there is a “hospital microbiome,” so “you can easily have that creep into your signal and mistake it for tumor-resident bacteria,” Dr. Vujkovic-Cvijin said.
Dr. Knight asserted that the samples are taken under sterile conditions, but other researchers do not think the level of sterility necessary for completely clean samples is possible.
“Just because it’s in your sample doesn’t mean it was in your tumor,” Dr. Gihawi said.
Even if scientists can retrieve a reliable sample without contamination, analyzing it requires comparing the genetic material to existing databases of microbial genomes. Yet, contamination and misclassification of genetic sequences can be problems in those reference genomes too, Dr. Gihawi explained.
Machine learning algorithms have a role in interpreting data, but “we need to be careful of what we use them for,” he added.
“These techniques are in their infancy, and we’re starting to chase them down, which is why we need to move microbiome research in a way that can be used clinically,” Dr. Gihawi said.
Influence on Cancer Treatment Outcomes
Again, however, the question of whether microbiomes exist within tumors is only one slice of the much larger field looking at microbiomes and cancer, including its influence on cancer treatment outcomes. Although much remains to be learned, less controversy exists over the thousands of studies in the past two decades that have gradually revealed how the body’s microbiome can affect both the course of a cancer and the effectiveness of different treatments.
The growing research showing the importance of the gut microbiome in cancer treatments is not surprising given its role in immunity more broadly. Because the human immune system must recognize and defend against microbes, the microbiome helps train it, Dr. Vujkovic-Cvijin said.
Some bacteria can escape the gut — a phenomenon called bacterial translocation — and may aid in fighting tumors. To grow large enough to be seen on imaging, tumors need to evolve several abilities, such as growing enough vascularization to receive blood flow and shutting down local immune responses.
“Any added boost, like immunotherapy, has a chance of breaking through that immune forcefield and killing the tumor cells,” Dr. Vujkovic-Cvijin said. Escaped gut bacteria may provide that boost.
“There’s a lot of evidence that depletion of the gut microbiome impairs immunotherapy and chemotherapy. The thinking behind some of those studies is that gut microbes can cross the gut barrier and when they do, they activate the immune system,” he said.
In mice engineered to have sterile guts, for example, the lack of bacteria results in less effective immune systems, Dr. Vujkovic-Cvijin pointed out. A host of research has shown that antibiotic exposure during and even 6 months before immunotherapy dramatically reduces survival rates. “That’s pretty convincing to me that gut microbes are important,” he said.
Dr. Vujkovic-Cvijin cautioned that there continues to be controversy on understanding which bacteria are important for response to immunotherapy. “The field is still in its infancy in terms of understanding which bacteria are most important for these effects,” he said.
Dr. Knight suggested that escaped bacteria may be the genesis of the ones that he and other researchers believe exist in tumors. “Because tumor microbes must come from somewhere, it is to be expected that some of those microbes will be co-opted from body-site specific commensals.”
It’s also possible that metabolites released from gut bacteria escape the gut and could theoretically affect distant tumor growth, Dr. Gihawi said. The most promising avenue of research in this area is metabolites being used as biomarkers, added Dr. Gihawi, whose lab published research on a link between bacteria detected in men’s urine and a more aggressive subset of prostate cancers. But that research is not far enough along to develop lab tests for clinical use, he noted.
No Consensus Yet
Even before the controversy erupted around Dr. Knight’s research, he co-founded the company Micronoma to develop cancer tests based on his microbe findings. The company has raised $17.5 million from private investors as of August 2023 and received the US Food and Drug Administration’s Breakthrough Device designation, allowing the firm to fast-track clinical trials testing the technology. The recent critiques have not changed the company’s plans.
It’s safe to say that scientists will continue to research and debate the possibility of tumor microbiomes until a consensus emerges.
“The field is evolving and studies testing the reproducibility of tumor-resident microbial signals are essential for developing our understanding in this area,” Dr. Vujkovic-Cvijin said.
Even if that path ultimately leads nowhere, as Dr. Salzberg expects, research into microbiomes and cancer has plenty of other directions to go.
“I’m actually quite an optimist,” Dr. Gihawi said. “I think there’s a lot of scope for some really good research here, especially in the sites where we know there is a strong microbiome, such as the gastrointestinal tract.”
A version of this article appeared on Medscape.com.
Last year, the controversy heightened when experts questioned a high-profile study — a 2020 analysis claiming that the tumors of 33 different cancers had their own unique microbiomes — on whether the “signature” of these bacterial compositions could help diagnose cancer.
The incident renewed the spotlight on “tumor microbiomes” because of the bold claims of the original paper and the strongly worded refutations of those claims. The broader field has focused primarily on ways the body’s microbiome interacts with cancers and cancer treatment.
This controversy has highlighted the challenges of making headway in a field where researchers may not even have the tools yet to puzzle-out the wide-ranging implications the microbiome holds for cancer diagnosis and treatment.
But it is also part of a provocative question within that larger field: whether tumors in the body, far from the natural microbiome in the gut, have their own thriving communities of bacteria, viruses, and fungi. And, if they do, how do those tumor microbiomes affect the development and progression of the cancer and the effectiveness of cancer therapies?
Cancer Controversy
The evidence is undeniable that some microbes can directly cause certain cancers and that the human gut microbiome can influence the effectiveness of certain therapies. Beyond that established science, however, the research has raised as many questions as answers about what we do and don’t know about microbiota and cancer.
The only confirmed microbiomes are on the skin and in the gut, mouth, and vagina, which are all areas with an easy direct route for bacteria to enter and grow in or on the body. A series of papers in recent years have suggested that other internal organs, and tumors within them, may have their own microbiomes.
“Whether microbes exist in tumors of internal organs beyond body surfaces exposed to the environment is a different matter,” said Ivan Vujkovic-Cvijin, PhD, an assistant professor of biomedical sciences and gastroenterology at Cedars-Sinai Medical Center in Los Angeles, whose lab studies how human gut microbes affect inflammatory diseases. “We’ve only recently had the tools to study that question on a molecular level, and the reported results have been conflicting.”
For example, research allegedly identified microbiota in the human placenta nearly one decade ago. But subsequent research contradicted those claims and showed that the source of the “placental microbiome” was actually contamination. Subsequent similar studies for other parts of the body faced the same scrutiny and, often, eventual debunking.
“Most likely, our immune system has undergone selective pressure to eliminate everything that crosses the gut barrier because there’s not much benefit to the body to have bacteria run amok in our internal organs,” Dr. Vujkovic-Cvijin said. “That can only disrupt the functioning of our tissues, to have an external organism living inside them.”
The controversy that erupted last summer, surrounding research from the lab of Rob Knight, PhD, at the University of California, San Diego, centered on a slightly different but related question: Could tumors harbor their own microbiomes?
This news organization spoke with two of the authors who published a paper contesting Dr. Knight’s findings: Steven Salzberg, PhD, a professor of biomedical engineering at John Hopkins Medicine, Baltimore, Maryland, and Abraham Gihawi, PhD, a research fellow at Norwich Medical School at the University of East Anglia in the United Kingdom.
Dr. Salzberg described two major problems with Dr. Knight’s study.
“What they found were false positives because of contamination in the database and flaws in their methods,” Dr. Salzberg said. “I can’t prove there’s no cancer microbiome, but I can say the cancer microbiomes that they reported don’t exist because the species they were finding aren’t there.”
Dr. Knight disagrees with Dr. Salzberg’s findings, noting that Dr. Salzberg and his co-authors did not examine the publicly available databases used in his study. In a written response, he said that his team’s examination of the database revealed that less than 1% of the microbial genomes overlapped with human ones and that removing them did not change their findings.
Dr. Knight also noted that his team could still “distinguish cancer types by their microbiome” even after running their analysis without the technique that Dr. Salzberg found fault with.
Dr. Salzberg said that the database linked above is not the one Dr. Knight’s study used, however. “The primary database in their study was never made public (it’s too large, they said), and it has/had about 69,000 genomes,” Dr. Salzberg said by email. “But even if we did, this is irrelevant. He’s trying to distract from the primary errors in their study,” which Dr. Salzberg said Dr. Knight’s team has not addressed.
The critiques Dr. Salzberg raised have been leveled at other studies investigating microbiomes specifically within tumors and independent of the body’s microbiome.
For example, a 2019 study in Nature described a fungal microbiome in pancreatic cancer that a Nature paper 4 years later directly contradicted, citing flaws that invalidated the original findings. A different 2019 study in Cell examined pancreatic tumor microbiota and patient outcomes, but it’s unclear whether the microorganisms moved from the gut to the pancreas or “constitute a durably colonized community that lives inside the tumor,” which remains a matter of debate, Dr. Vujkovic-Cvijin said.
A 2020 study in Science suggested diverse microbial communities in seven tumor types, but those findings were similarly called into question. That study stated that “bacteria were first detected in human tumors more than 100 years ago” and that “bacteria are well-known residents in human tumors,” but Dr. Salzberg considers those statements misleading.
It’s true that bacteria and viruses have been detected in tumors because “there’s very good evidence that an acute infection caused by a very small number of viruses and bacteria can cause a tumor,” Dr. Salzberg said. Human papillomavirus, for example, can cause six different types of cancer. Inflammation and ulcers caused by Helicobacter pylori may progress to stomach cancer, and Fusobacterium nucleatum and Enterococcus faecalis have been shown to contribute to colorectal cancer. Those examples differ from a microbiome; this “a community of bacteria and possibly other microscopic bugs, like fungi, that are happily living in the tumor” the same way microbes reside in our guts, he said.
Dr. Knight said that many bacteria his team identified “have been confirmed independently in subsequent work.” He acknowledged, however, that more research is needed.
Several of the contested studies above were among a lengthy list that Dr. Knight provided, noting that most of the disagreements “have two sides to them, and critiques from one particular group does not immediately invalidate a reported finding.”
Yet, many of the papers Dr. Knight listed are precisely the types that skeptics like Dr. Salzberg believe are too flawed to draw reliable conclusions.
“I think many agree that microbes may exist within tumors that are exposed to the environment, like tumors of the skin, gut, and mouth,” Dr. Vujkovic-Cvijin said. It’s less clear, however, whether tumors further from the body’s microbiome harbor any microbes or where they came from if they do. Microbial signals in organs elsewhere in the body become faint quickly, he said.
Underdeveloped Technology
Though Dr. Salzberg said that the concept of a tumor microbiome is “implausible” because there’s no easy route for bacteria to reach internal organs, it’s unclear whether scientists have the technology yet to adequately answer this question.
For one thing, samples in these types of studies are typically “ultra-low biomass samples, where the signal — the amount of microbes in the sample — is so low that it’s comparable to how much would be expected to be found in reagents and environmental contamination through processing,” Dr. Vujkovic-Cvijin explained. Many polymerases used to amplify a DNA signal, for example, are made in bacteria and may retain trace amounts identified in these studies.
Dr. Knight agreed that low biomass is a challenge in this field but is not an unsurmountable one.
Another challenge is that study samples, as with Dr. Knight’s work, were collected during routine surgeries without the intent to find a microbial signal. Simply using a scalpel to cut through the skin means cutting through a layer of bacteria, and surgery rooms are not designed to eliminate all bacteria. Some work has even shown there is a “hospital microbiome,” so “you can easily have that creep into your signal and mistake it for tumor-resident bacteria,” Dr. Vujkovic-Cvijin said.
Dr. Knight asserted that the samples are taken under sterile conditions, but other researchers do not think the level of sterility necessary for completely clean samples is possible.
“Just because it’s in your sample doesn’t mean it was in your tumor,” Dr. Gihawi said.
Even if scientists can retrieve a reliable sample without contamination, analyzing it requires comparing the genetic material to existing databases of microbial genomes. Yet, contamination and misclassification of genetic sequences can be problems in those reference genomes too, Dr. Gihawi explained.
Machine learning algorithms have a role in interpreting data, but “we need to be careful of what we use them for,” he added.
“These techniques are in their infancy, and we’re starting to chase them down, which is why we need to move microbiome research in a way that can be used clinically,” Dr. Gihawi said.
Influence on Cancer Treatment Outcomes
Again, however, the question of whether microbiomes exist within tumors is only one slice of the much larger field looking at microbiomes and cancer, including its influence on cancer treatment outcomes. Although much remains to be learned, less controversy exists over the thousands of studies in the past two decades that have gradually revealed how the body’s microbiome can affect both the course of a cancer and the effectiveness of different treatments.
The growing research showing the importance of the gut microbiome in cancer treatments is not surprising given its role in immunity more broadly. Because the human immune system must recognize and defend against microbes, the microbiome helps train it, Dr. Vujkovic-Cvijin said.
Some bacteria can escape the gut — a phenomenon called bacterial translocation — and may aid in fighting tumors. To grow large enough to be seen on imaging, tumors need to evolve several abilities, such as growing enough vascularization to receive blood flow and shutting down local immune responses.
“Any added boost, like immunotherapy, has a chance of breaking through that immune forcefield and killing the tumor cells,” Dr. Vujkovic-Cvijin said. Escaped gut bacteria may provide that boost.
“There’s a lot of evidence that depletion of the gut microbiome impairs immunotherapy and chemotherapy. The thinking behind some of those studies is that gut microbes can cross the gut barrier and when they do, they activate the immune system,” he said.
In mice engineered to have sterile guts, for example, the lack of bacteria results in less effective immune systems, Dr. Vujkovic-Cvijin pointed out. A host of research has shown that antibiotic exposure during and even 6 months before immunotherapy dramatically reduces survival rates. “That’s pretty convincing to me that gut microbes are important,” he said.
Dr. Vujkovic-Cvijin cautioned that there continues to be controversy on understanding which bacteria are important for response to immunotherapy. “The field is still in its infancy in terms of understanding which bacteria are most important for these effects,” he said.
Dr. Knight suggested that escaped bacteria may be the genesis of the ones that he and other researchers believe exist in tumors. “Because tumor microbes must come from somewhere, it is to be expected that some of those microbes will be co-opted from body-site specific commensals.”
It’s also possible that metabolites released from gut bacteria escape the gut and could theoretically affect distant tumor growth, Dr. Gihawi said. The most promising avenue of research in this area is metabolites being used as biomarkers, added Dr. Gihawi, whose lab published research on a link between bacteria detected in men’s urine and a more aggressive subset of prostate cancers. But that research is not far enough along to develop lab tests for clinical use, he noted.
No Consensus Yet
Even before the controversy erupted around Dr. Knight’s research, he co-founded the company Micronoma to develop cancer tests based on his microbe findings. The company has raised $17.5 million from private investors as of August 2023 and received the US Food and Drug Administration’s Breakthrough Device designation, allowing the firm to fast-track clinical trials testing the technology. The recent critiques have not changed the company’s plans.
It’s safe to say that scientists will continue to research and debate the possibility of tumor microbiomes until a consensus emerges.
“The field is evolving and studies testing the reproducibility of tumor-resident microbial signals are essential for developing our understanding in this area,” Dr. Vujkovic-Cvijin said.
Even if that path ultimately leads nowhere, as Dr. Salzberg expects, research into microbiomes and cancer has plenty of other directions to go.
“I’m actually quite an optimist,” Dr. Gihawi said. “I think there’s a lot of scope for some really good research here, especially in the sites where we know there is a strong microbiome, such as the gastrointestinal tract.”
A version of this article appeared on Medscape.com.
Last year, the controversy heightened when experts questioned a high-profile study — a 2020 analysis claiming that the tumors of 33 different cancers had their own unique microbiomes — on whether the “signature” of these bacterial compositions could help diagnose cancer.
The incident renewed the spotlight on “tumor microbiomes” because of the bold claims of the original paper and the strongly worded refutations of those claims. The broader field has focused primarily on ways the body’s microbiome interacts with cancers and cancer treatment.
This controversy has highlighted the challenges of making headway in a field where researchers may not even have the tools yet to puzzle-out the wide-ranging implications the microbiome holds for cancer diagnosis and treatment.
But it is also part of a provocative question within that larger field: whether tumors in the body, far from the natural microbiome in the gut, have their own thriving communities of bacteria, viruses, and fungi. And, if they do, how do those tumor microbiomes affect the development and progression of the cancer and the effectiveness of cancer therapies?
Cancer Controversy
The evidence is undeniable that some microbes can directly cause certain cancers and that the human gut microbiome can influence the effectiveness of certain therapies. Beyond that established science, however, the research has raised as many questions as answers about what we do and don’t know about microbiota and cancer.
The only confirmed microbiomes are on the skin and in the gut, mouth, and vagina, which are all areas with an easy direct route for bacteria to enter and grow in or on the body. A series of papers in recent years have suggested that other internal organs, and tumors within them, may have their own microbiomes.
“Whether microbes exist in tumors of internal organs beyond body surfaces exposed to the environment is a different matter,” said Ivan Vujkovic-Cvijin, PhD, an assistant professor of biomedical sciences and gastroenterology at Cedars-Sinai Medical Center in Los Angeles, whose lab studies how human gut microbes affect inflammatory diseases. “We’ve only recently had the tools to study that question on a molecular level, and the reported results have been conflicting.”
For example, research allegedly identified microbiota in the human placenta nearly one decade ago. But subsequent research contradicted those claims and showed that the source of the “placental microbiome” was actually contamination. Subsequent similar studies for other parts of the body faced the same scrutiny and, often, eventual debunking.
“Most likely, our immune system has undergone selective pressure to eliminate everything that crosses the gut barrier because there’s not much benefit to the body to have bacteria run amok in our internal organs,” Dr. Vujkovic-Cvijin said. “That can only disrupt the functioning of our tissues, to have an external organism living inside them.”
The controversy that erupted last summer, surrounding research from the lab of Rob Knight, PhD, at the University of California, San Diego, centered on a slightly different but related question: Could tumors harbor their own microbiomes?
This news organization spoke with two of the authors who published a paper contesting Dr. Knight’s findings: Steven Salzberg, PhD, a professor of biomedical engineering at John Hopkins Medicine, Baltimore, Maryland, and Abraham Gihawi, PhD, a research fellow at Norwich Medical School at the University of East Anglia in the United Kingdom.
Dr. Salzberg described two major problems with Dr. Knight’s study.
“What they found were false positives because of contamination in the database and flaws in their methods,” Dr. Salzberg said. “I can’t prove there’s no cancer microbiome, but I can say the cancer microbiomes that they reported don’t exist because the species they were finding aren’t there.”
Dr. Knight disagrees with Dr. Salzberg’s findings, noting that Dr. Salzberg and his co-authors did not examine the publicly available databases used in his study. In a written response, he said that his team’s examination of the database revealed that less than 1% of the microbial genomes overlapped with human ones and that removing them did not change their findings.
Dr. Knight also noted that his team could still “distinguish cancer types by their microbiome” even after running their analysis without the technique that Dr. Salzberg found fault with.
Dr. Salzberg said that the database linked above is not the one Dr. Knight’s study used, however. “The primary database in their study was never made public (it’s too large, they said), and it has/had about 69,000 genomes,” Dr. Salzberg said by email. “But even if we did, this is irrelevant. He’s trying to distract from the primary errors in their study,” which Dr. Salzberg said Dr. Knight’s team has not addressed.
The critiques Dr. Salzberg raised have been leveled at other studies investigating microbiomes specifically within tumors and independent of the body’s microbiome.
For example, a 2019 study in Nature described a fungal microbiome in pancreatic cancer that a Nature paper 4 years later directly contradicted, citing flaws that invalidated the original findings. A different 2019 study in Cell examined pancreatic tumor microbiota and patient outcomes, but it’s unclear whether the microorganisms moved from the gut to the pancreas or “constitute a durably colonized community that lives inside the tumor,” which remains a matter of debate, Dr. Vujkovic-Cvijin said.
A 2020 study in Science suggested diverse microbial communities in seven tumor types, but those findings were similarly called into question. That study stated that “bacteria were first detected in human tumors more than 100 years ago” and that “bacteria are well-known residents in human tumors,” but Dr. Salzberg considers those statements misleading.
It’s true that bacteria and viruses have been detected in tumors because “there’s very good evidence that an acute infection caused by a very small number of viruses and bacteria can cause a tumor,” Dr. Salzberg said. Human papillomavirus, for example, can cause six different types of cancer. Inflammation and ulcers caused by Helicobacter pylori may progress to stomach cancer, and Fusobacterium nucleatum and Enterococcus faecalis have been shown to contribute to colorectal cancer. Those examples differ from a microbiome; this “a community of bacteria and possibly other microscopic bugs, like fungi, that are happily living in the tumor” the same way microbes reside in our guts, he said.
Dr. Knight said that many bacteria his team identified “have been confirmed independently in subsequent work.” He acknowledged, however, that more research is needed.
Several of the contested studies above were among a lengthy list that Dr. Knight provided, noting that most of the disagreements “have two sides to them, and critiques from one particular group does not immediately invalidate a reported finding.”
Yet, many of the papers Dr. Knight listed are precisely the types that skeptics like Dr. Salzberg believe are too flawed to draw reliable conclusions.
“I think many agree that microbes may exist within tumors that are exposed to the environment, like tumors of the skin, gut, and mouth,” Dr. Vujkovic-Cvijin said. It’s less clear, however, whether tumors further from the body’s microbiome harbor any microbes or where they came from if they do. Microbial signals in organs elsewhere in the body become faint quickly, he said.
Underdeveloped Technology
Though Dr. Salzberg said that the concept of a tumor microbiome is “implausible” because there’s no easy route for bacteria to reach internal organs, it’s unclear whether scientists have the technology yet to adequately answer this question.
For one thing, samples in these types of studies are typically “ultra-low biomass samples, where the signal — the amount of microbes in the sample — is so low that it’s comparable to how much would be expected to be found in reagents and environmental contamination through processing,” Dr. Vujkovic-Cvijin explained. Many polymerases used to amplify a DNA signal, for example, are made in bacteria and may retain trace amounts identified in these studies.
Dr. Knight agreed that low biomass is a challenge in this field but is not an unsurmountable one.
Another challenge is that study samples, as with Dr. Knight’s work, were collected during routine surgeries without the intent to find a microbial signal. Simply using a scalpel to cut through the skin means cutting through a layer of bacteria, and surgery rooms are not designed to eliminate all bacteria. Some work has even shown there is a “hospital microbiome,” so “you can easily have that creep into your signal and mistake it for tumor-resident bacteria,” Dr. Vujkovic-Cvijin said.
Dr. Knight asserted that the samples are taken under sterile conditions, but other researchers do not think the level of sterility necessary for completely clean samples is possible.
“Just because it’s in your sample doesn’t mean it was in your tumor,” Dr. Gihawi said.
Even if scientists can retrieve a reliable sample without contamination, analyzing it requires comparing the genetic material to existing databases of microbial genomes. Yet, contamination and misclassification of genetic sequences can be problems in those reference genomes too, Dr. Gihawi explained.
Machine learning algorithms have a role in interpreting data, but “we need to be careful of what we use them for,” he added.
“These techniques are in their infancy, and we’re starting to chase them down, which is why we need to move microbiome research in a way that can be used clinically,” Dr. Gihawi said.
Influence on Cancer Treatment Outcomes
Again, however, the question of whether microbiomes exist within tumors is only one slice of the much larger field looking at microbiomes and cancer, including its influence on cancer treatment outcomes. Although much remains to be learned, less controversy exists over the thousands of studies in the past two decades that have gradually revealed how the body’s microbiome can affect both the course of a cancer and the effectiveness of different treatments.
The growing research showing the importance of the gut microbiome in cancer treatments is not surprising given its role in immunity more broadly. Because the human immune system must recognize and defend against microbes, the microbiome helps train it, Dr. Vujkovic-Cvijin said.
Some bacteria can escape the gut — a phenomenon called bacterial translocation — and may aid in fighting tumors. To grow large enough to be seen on imaging, tumors need to evolve several abilities, such as growing enough vascularization to receive blood flow and shutting down local immune responses.
“Any added boost, like immunotherapy, has a chance of breaking through that immune forcefield and killing the tumor cells,” Dr. Vujkovic-Cvijin said. Escaped gut bacteria may provide that boost.
“There’s a lot of evidence that depletion of the gut microbiome impairs immunotherapy and chemotherapy. The thinking behind some of those studies is that gut microbes can cross the gut barrier and when they do, they activate the immune system,” he said.
In mice engineered to have sterile guts, for example, the lack of bacteria results in less effective immune systems, Dr. Vujkovic-Cvijin pointed out. A host of research has shown that antibiotic exposure during and even 6 months before immunotherapy dramatically reduces survival rates. “That’s pretty convincing to me that gut microbes are important,” he said.
Dr. Vujkovic-Cvijin cautioned that there continues to be controversy on understanding which bacteria are important for response to immunotherapy. “The field is still in its infancy in terms of understanding which bacteria are most important for these effects,” he said.
Dr. Knight suggested that escaped bacteria may be the genesis of the ones that he and other researchers believe exist in tumors. “Because tumor microbes must come from somewhere, it is to be expected that some of those microbes will be co-opted from body-site specific commensals.”
It’s also possible that metabolites released from gut bacteria escape the gut and could theoretically affect distant tumor growth, Dr. Gihawi said. The most promising avenue of research in this area is metabolites being used as biomarkers, added Dr. Gihawi, whose lab published research on a link between bacteria detected in men’s urine and a more aggressive subset of prostate cancers. But that research is not far enough along to develop lab tests for clinical use, he noted.
No Consensus Yet
Even before the controversy erupted around Dr. Knight’s research, he co-founded the company Micronoma to develop cancer tests based on his microbe findings. The company has raised $17.5 million from private investors as of August 2023 and received the US Food and Drug Administration’s Breakthrough Device designation, allowing the firm to fast-track clinical trials testing the technology. The recent critiques have not changed the company’s plans.
It’s safe to say that scientists will continue to research and debate the possibility of tumor microbiomes until a consensus emerges.
“The field is evolving and studies testing the reproducibility of tumor-resident microbial signals are essential for developing our understanding in this area,” Dr. Vujkovic-Cvijin said.
Even if that path ultimately leads nowhere, as Dr. Salzberg expects, research into microbiomes and cancer has plenty of other directions to go.
“I’m actually quite an optimist,” Dr. Gihawi said. “I think there’s a lot of scope for some really good research here, especially in the sites where we know there is a strong microbiome, such as the gastrointestinal tract.”
A version of this article appeared on Medscape.com.
Few Cancer Survivors Meet ACS Nutrition, Exercise Guidelines
TOPLINE:
METHODOLOGY:
- The ACS has published nutrition and exercise guidelines for cancer survivors, which include recommendations to maintain a healthy weight and diet, cut out alcohol, and participate in regular physical activities. Engaging in these behaviors is associated with longer survival among cancer survivors, but whether survivors follow these nutrition and activity recommendations has not been systematically tracked.
- Researchers evaluated data on 10,020 individuals (mean age, 64.2 years) who had completed cancer treatment. Data came from the Behavioral Risk Factor Surveillance System telephone-based survey administered in 2017, 2019, and 2021, which represents 2.7 million cancer survivors.
- The researchers estimated survivors’ adherence to guidelines across four domains: Weight, physical activity, fruit and vegetable consumption, and alcohol intake. Factors associated with adherence were also evaluated.
- Overall, 9,121 survivors (91%) completed questionnaires for all four domains.
TAKEAWAY:
Only 4% of patients (365 of 9121) followed ACS guidelines in all four categories.
When assessing adherence to each category, the researchers found that 72% of cancer survivors reported engaging in recommended levels of physical activity, 68% maintained a nonobese weight, 50% said they did not consume alcohol, and 12% said they consumed recommended quantities of fruits and vegetables.
Compared with people in the general population, cancer survivors generally engaged in fewer healthy behaviors than those who had never been diagnosed with cancer.
The authors identified certain factors associated with greater guideline adherence, including female sex, older age, Black (vs White) race, and higher education level (college graduate).
IN PRACTICE:
This study highlights a potential “gap between published guidelines regarding behavioral modifications for cancer survivors and uptake of these behaviors,” the authors wrote, adding that “it is essential for oncologists and general internists to improve widespread and systematic counseling on these guidelines to improve uptake of healthy behaviors in this vulnerable patient population.”
SOURCE:
This work, led by Carter Baughman, MD, from the Division of Internal Medicine at Beth Israel Deaconess Medical Center, Boston, Massachusetts, was published online in JAMA Oncology.
LIMITATIONS:
The authors reported several study limitations, most notably that self-reported data may introduce biases.
DISCLOSURES:
The study funding source was not reported. One author received grants from the US Highbush Blueberry Council outside the submitted work. No other disclosures were reported.
A version of this article appeared on Medscape.com.
TOPLINE:
METHODOLOGY:
- The ACS has published nutrition and exercise guidelines for cancer survivors, which include recommendations to maintain a healthy weight and diet, cut out alcohol, and participate in regular physical activities. Engaging in these behaviors is associated with longer survival among cancer survivors, but whether survivors follow these nutrition and activity recommendations has not been systematically tracked.
- Researchers evaluated data on 10,020 individuals (mean age, 64.2 years) who had completed cancer treatment. Data came from the Behavioral Risk Factor Surveillance System telephone-based survey administered in 2017, 2019, and 2021, which represents 2.7 million cancer survivors.
- The researchers estimated survivors’ adherence to guidelines across four domains: Weight, physical activity, fruit and vegetable consumption, and alcohol intake. Factors associated with adherence were also evaluated.
- Overall, 9,121 survivors (91%) completed questionnaires for all four domains.
TAKEAWAY:
Only 4% of patients (365 of 9121) followed ACS guidelines in all four categories.
When assessing adherence to each category, the researchers found that 72% of cancer survivors reported engaging in recommended levels of physical activity, 68% maintained a nonobese weight, 50% said they did not consume alcohol, and 12% said they consumed recommended quantities of fruits and vegetables.
Compared with people in the general population, cancer survivors generally engaged in fewer healthy behaviors than those who had never been diagnosed with cancer.
The authors identified certain factors associated with greater guideline adherence, including female sex, older age, Black (vs White) race, and higher education level (college graduate).
IN PRACTICE:
This study highlights a potential “gap between published guidelines regarding behavioral modifications for cancer survivors and uptake of these behaviors,” the authors wrote, adding that “it is essential for oncologists and general internists to improve widespread and systematic counseling on these guidelines to improve uptake of healthy behaviors in this vulnerable patient population.”
SOURCE:
This work, led by Carter Baughman, MD, from the Division of Internal Medicine at Beth Israel Deaconess Medical Center, Boston, Massachusetts, was published online in JAMA Oncology.
LIMITATIONS:
The authors reported several study limitations, most notably that self-reported data may introduce biases.
DISCLOSURES:
The study funding source was not reported. One author received grants from the US Highbush Blueberry Council outside the submitted work. No other disclosures were reported.
A version of this article appeared on Medscape.com.
TOPLINE:
METHODOLOGY:
- The ACS has published nutrition and exercise guidelines for cancer survivors, which include recommendations to maintain a healthy weight and diet, cut out alcohol, and participate in regular physical activities. Engaging in these behaviors is associated with longer survival among cancer survivors, but whether survivors follow these nutrition and activity recommendations has not been systematically tracked.
- Researchers evaluated data on 10,020 individuals (mean age, 64.2 years) who had completed cancer treatment. Data came from the Behavioral Risk Factor Surveillance System telephone-based survey administered in 2017, 2019, and 2021, which represents 2.7 million cancer survivors.
- The researchers estimated survivors’ adherence to guidelines across four domains: Weight, physical activity, fruit and vegetable consumption, and alcohol intake. Factors associated with adherence were also evaluated.
- Overall, 9,121 survivors (91%) completed questionnaires for all four domains.
TAKEAWAY:
Only 4% of patients (365 of 9121) followed ACS guidelines in all four categories.
When assessing adherence to each category, the researchers found that 72% of cancer survivors reported engaging in recommended levels of physical activity, 68% maintained a nonobese weight, 50% said they did not consume alcohol, and 12% said they consumed recommended quantities of fruits and vegetables.
Compared with people in the general population, cancer survivors generally engaged in fewer healthy behaviors than those who had never been diagnosed with cancer.
The authors identified certain factors associated with greater guideline adherence, including female sex, older age, Black (vs White) race, and higher education level (college graduate).
IN PRACTICE:
This study highlights a potential “gap between published guidelines regarding behavioral modifications for cancer survivors and uptake of these behaviors,” the authors wrote, adding that “it is essential for oncologists and general internists to improve widespread and systematic counseling on these guidelines to improve uptake of healthy behaviors in this vulnerable patient population.”
SOURCE:
This work, led by Carter Baughman, MD, from the Division of Internal Medicine at Beth Israel Deaconess Medical Center, Boston, Massachusetts, was published online in JAMA Oncology.
LIMITATIONS:
The authors reported several study limitations, most notably that self-reported data may introduce biases.
DISCLOSURES:
The study funding source was not reported. One author received grants from the US Highbush Blueberry Council outside the submitted work. No other disclosures were reported.
A version of this article appeared on Medscape.com.
Keratoacanthoma, SCC Relatively Rare With PD-1/PD-L1 Inhibitors, Study Suggests
TOPLINE:
(AEs) reported to the US Food and Drug Administration (FDA).
METHODOLOGY:
- The risk for dermatologic immune-related side effects may be increased with immunologic-modifying drugs.
- To determine if there are significant signals between keratoacanthomas and cSCCs and PD-1/PD-L1 inhibitors, researchers analyzed AEs associated with these agents reported to the FDA’s Adverse Event Reporting System (FAERS) between January 2004 and May 2023.
- Pharmacovigilance signals were identified, and a significant signal was defined as the lower 95% CI of a reporting odds ratio (ROR) greater than one or the lower 95% CI of an information component (IC) greater than 0.
TAKEAWAY:
- Of the 158,000 reports of PD-1/PD-L1 inhibitor use, 43 were in patients who developed a keratoacanthoma (mean age, 77 years; 39% women) and 83 were in patients who developed cSCC (mean age, 71 years; 41% women). Patients aged 60-79 years were most likely to develop keratoacanthomas and cSCC on these treatments.
- A PD-1/PD-L1 inhibitor was listed as the suspect drug in all 43 keratoacanthoma reports and in 70 of 83 cSCC reports (the remaining 13 listed them as the concomitant drug).
- Significant signals were reported for both keratoacanthoma (ROR, 9.7; IC, 1.9) and cSCC (ROR, 3.0; IC, 0.9) with PD-1/PD-L1 inhibitor use.
- Of the reports where this information was available, all 10 cases of PD-1/PD-L1 inhibitor–linked keratoacanthoma and 10 of 17 cases (59%) of PD-1/PD-L1 inhibitor–linked cSCC, resolution was noted following discontinuation or dose reduction of the inhibitor.
IN PRACTICE:
“Given the large number of patients receiving immunotherapy, FAERS recording only 43 patients developing keratoacanthoma and 83 patients developing cSCC highlights that these conditions are relatively rare adverse events,” the authors wrote but added that more studies are needed to confirm these results.
SOURCE:
The study, led by Pushkar Aggarwal, MD, MBA, of the Department of Dermatology, University of Cincinnati, Cincinnati, Ohio, was published online in JAMA Dermatology.
LIMITATIONS:
The data obtained from FAERS did not contain information on all AEs from drugs. In addition, a causal association could not be determined.
DISCLOSURES:
The funding source was not reported. The authors did not report any conflicts of interest.
A version of this article appeared on Medscape.com.
TOPLINE:
(AEs) reported to the US Food and Drug Administration (FDA).
METHODOLOGY:
- The risk for dermatologic immune-related side effects may be increased with immunologic-modifying drugs.
- To determine if there are significant signals between keratoacanthomas and cSCCs and PD-1/PD-L1 inhibitors, researchers analyzed AEs associated with these agents reported to the FDA’s Adverse Event Reporting System (FAERS) between January 2004 and May 2023.
- Pharmacovigilance signals were identified, and a significant signal was defined as the lower 95% CI of a reporting odds ratio (ROR) greater than one or the lower 95% CI of an information component (IC) greater than 0.
TAKEAWAY:
- Of the 158,000 reports of PD-1/PD-L1 inhibitor use, 43 were in patients who developed a keratoacanthoma (mean age, 77 years; 39% women) and 83 were in patients who developed cSCC (mean age, 71 years; 41% women). Patients aged 60-79 years were most likely to develop keratoacanthomas and cSCC on these treatments.
- A PD-1/PD-L1 inhibitor was listed as the suspect drug in all 43 keratoacanthoma reports and in 70 of 83 cSCC reports (the remaining 13 listed them as the concomitant drug).
- Significant signals were reported for both keratoacanthoma (ROR, 9.7; IC, 1.9) and cSCC (ROR, 3.0; IC, 0.9) with PD-1/PD-L1 inhibitor use.
- Of the reports where this information was available, all 10 cases of PD-1/PD-L1 inhibitor–linked keratoacanthoma and 10 of 17 cases (59%) of PD-1/PD-L1 inhibitor–linked cSCC, resolution was noted following discontinuation or dose reduction of the inhibitor.
IN PRACTICE:
“Given the large number of patients receiving immunotherapy, FAERS recording only 43 patients developing keratoacanthoma and 83 patients developing cSCC highlights that these conditions are relatively rare adverse events,” the authors wrote but added that more studies are needed to confirm these results.
SOURCE:
The study, led by Pushkar Aggarwal, MD, MBA, of the Department of Dermatology, University of Cincinnati, Cincinnati, Ohio, was published online in JAMA Dermatology.
LIMITATIONS:
The data obtained from FAERS did not contain information on all AEs from drugs. In addition, a causal association could not be determined.
DISCLOSURES:
The funding source was not reported. The authors did not report any conflicts of interest.
A version of this article appeared on Medscape.com.
TOPLINE:
(AEs) reported to the US Food and Drug Administration (FDA).
METHODOLOGY:
- The risk for dermatologic immune-related side effects may be increased with immunologic-modifying drugs.
- To determine if there are significant signals between keratoacanthomas and cSCCs and PD-1/PD-L1 inhibitors, researchers analyzed AEs associated with these agents reported to the FDA’s Adverse Event Reporting System (FAERS) between January 2004 and May 2023.
- Pharmacovigilance signals were identified, and a significant signal was defined as the lower 95% CI of a reporting odds ratio (ROR) greater than one or the lower 95% CI of an information component (IC) greater than 0.
TAKEAWAY:
- Of the 158,000 reports of PD-1/PD-L1 inhibitor use, 43 were in patients who developed a keratoacanthoma (mean age, 77 years; 39% women) and 83 were in patients who developed cSCC (mean age, 71 years; 41% women). Patients aged 60-79 years were most likely to develop keratoacanthomas and cSCC on these treatments.
- A PD-1/PD-L1 inhibitor was listed as the suspect drug in all 43 keratoacanthoma reports and in 70 of 83 cSCC reports (the remaining 13 listed them as the concomitant drug).
- Significant signals were reported for both keratoacanthoma (ROR, 9.7; IC, 1.9) and cSCC (ROR, 3.0; IC, 0.9) with PD-1/PD-L1 inhibitor use.
- Of the reports where this information was available, all 10 cases of PD-1/PD-L1 inhibitor–linked keratoacanthoma and 10 of 17 cases (59%) of PD-1/PD-L1 inhibitor–linked cSCC, resolution was noted following discontinuation or dose reduction of the inhibitor.
IN PRACTICE:
“Given the large number of patients receiving immunotherapy, FAERS recording only 43 patients developing keratoacanthoma and 83 patients developing cSCC highlights that these conditions are relatively rare adverse events,” the authors wrote but added that more studies are needed to confirm these results.
SOURCE:
The study, led by Pushkar Aggarwal, MD, MBA, of the Department of Dermatology, University of Cincinnati, Cincinnati, Ohio, was published online in JAMA Dermatology.
LIMITATIONS:
The data obtained from FAERS did not contain information on all AEs from drugs. In addition, a causal association could not be determined.
DISCLOSURES:
The funding source was not reported. The authors did not report any conflicts of interest.
A version of this article appeared on Medscape.com.
Most Targeted Cancer Drugs Lack Substantial Clinical Benefit
TOPLINE:
METHODOLOGY:
- The strength and quality of evidence supporting genome-targeted cancer drug approvals vary. A big reason is the growing number of cancer drug approvals based on surrogate endpoints, such as disease-free and progression-free survival, instead of clinical endpoints, such as overall survival or quality of life. The US Food and Drug Administration (FDA) has also approved genome-targeted cancer drugs based on phase 1 or single-arm trials.
- Given these less rigorous considerations for approval, “the validity and value of the targets and surrogate measures underlying FDA genome-targeted cancer drug approvals are uncertain,” the researchers explained.
- In the current analysis, researchers assessed the validity of the molecular targets as well as the clinical benefits of genome-targeted cancer drugs approved in the United States from 2015 to 2022 based on results from pivotal trials.
- The researchers evaluated the strength of evidence supporting molecular targetability using the European Society for Medical Oncology (ESMO) Scale for Clinical Actionability of Molecular Targets (ESCAT) and the clinical benefit using the ESMO–Magnitude of Clinical Benefit Scale (ESMO-MCBS).
- The authors defined a substantial clinical benefit as an A or B grade for curative intent and a 4 or 5 for noncurative intent. High-benefit genomic-based cancer treatments were defined as those associated with a substantial clinical benefit (ESMO-MCBS) and that qualified as ESCAT category level I-A (a clinical benefit based on prospective randomized data) or I-B (prospective nonrandomized data).
TAKEAWAY:
- The analyses focused on 50 molecular-targeted cancer drugs covering 84 indications. Of which, 45 indications (54%) were approved based on phase 1 or 2 pivotal trials, 45 (54%) were supported by single-arm pivotal trials and the remaining 39 (46%) by randomized trial, and 48 (57%) were approved based on subgroup analyses.
- Among the 84 indications, more than half (55%) of the pivotal trials supporting approval used overall response rate as a primary endpoint, 31% used progression-free survival, and 6% used disease-free survival. Only seven indications (8%) were supported by pivotal trials demonstrating an improvement in overall survival.
- Among the 84 trials, 24 (29%) met the ESMO-MCBS threshold for substantial clinical benefit.
- Overall, when combining all ratings, only 24 of the 84 indications (29%) were considered high-benefit genomic-based cancer treatments.
IN PRACTICE:
“We applied the ESMO-MCBS and ESCAT value frameworks to identify therapies and molecular targets providing high clinical value that should be widely available to patients” and “found that drug indications supported by these characteristics represent a minority of cancer drug approvals in recent years,” the authors said. Using these value frameworks could help payers, governments, and individual patients “prioritize the availability of high-value molecular-targeted therapies.”
SOURCE:
The study, with first author Ariadna Tibau, MD, PhD, Brigham and Women’s Hospital and Harvard Medical School, Boston, was published online in JAMA Oncology.
LIMITATIONS:
The study evaluated only trials that supported regulatory approval and did not include outcomes of postapproval clinical studies, which could lead to changes in ESMO-MCBS grades and ESCAT levels of evidence over time.
DISCLOSURES:
The study was funded by the Kaiser Permanente Institute for Health Policy, Arnold Ventures, and the Commonwealth Fund. The authors had no relevant disclosures.
A version of this article appeared on Medscape.com.
TOPLINE:
METHODOLOGY:
- The strength and quality of evidence supporting genome-targeted cancer drug approvals vary. A big reason is the growing number of cancer drug approvals based on surrogate endpoints, such as disease-free and progression-free survival, instead of clinical endpoints, such as overall survival or quality of life. The US Food and Drug Administration (FDA) has also approved genome-targeted cancer drugs based on phase 1 or single-arm trials.
- Given these less rigorous considerations for approval, “the validity and value of the targets and surrogate measures underlying FDA genome-targeted cancer drug approvals are uncertain,” the researchers explained.
- In the current analysis, researchers assessed the validity of the molecular targets as well as the clinical benefits of genome-targeted cancer drugs approved in the United States from 2015 to 2022 based on results from pivotal trials.
- The researchers evaluated the strength of evidence supporting molecular targetability using the European Society for Medical Oncology (ESMO) Scale for Clinical Actionability of Molecular Targets (ESCAT) and the clinical benefit using the ESMO–Magnitude of Clinical Benefit Scale (ESMO-MCBS).
- The authors defined a substantial clinical benefit as an A or B grade for curative intent and a 4 or 5 for noncurative intent. High-benefit genomic-based cancer treatments were defined as those associated with a substantial clinical benefit (ESMO-MCBS) and that qualified as ESCAT category level I-A (a clinical benefit based on prospective randomized data) or I-B (prospective nonrandomized data).
TAKEAWAY:
- The analyses focused on 50 molecular-targeted cancer drugs covering 84 indications. Of which, 45 indications (54%) were approved based on phase 1 or 2 pivotal trials, 45 (54%) were supported by single-arm pivotal trials and the remaining 39 (46%) by randomized trial, and 48 (57%) were approved based on subgroup analyses.
- Among the 84 indications, more than half (55%) of the pivotal trials supporting approval used overall response rate as a primary endpoint, 31% used progression-free survival, and 6% used disease-free survival. Only seven indications (8%) were supported by pivotal trials demonstrating an improvement in overall survival.
- Among the 84 trials, 24 (29%) met the ESMO-MCBS threshold for substantial clinical benefit.
- Overall, when combining all ratings, only 24 of the 84 indications (29%) were considered high-benefit genomic-based cancer treatments.
IN PRACTICE:
“We applied the ESMO-MCBS and ESCAT value frameworks to identify therapies and molecular targets providing high clinical value that should be widely available to patients” and “found that drug indications supported by these characteristics represent a minority of cancer drug approvals in recent years,” the authors said. Using these value frameworks could help payers, governments, and individual patients “prioritize the availability of high-value molecular-targeted therapies.”
SOURCE:
The study, with first author Ariadna Tibau, MD, PhD, Brigham and Women’s Hospital and Harvard Medical School, Boston, was published online in JAMA Oncology.
LIMITATIONS:
The study evaluated only trials that supported regulatory approval and did not include outcomes of postapproval clinical studies, which could lead to changes in ESMO-MCBS grades and ESCAT levels of evidence over time.
DISCLOSURES:
The study was funded by the Kaiser Permanente Institute for Health Policy, Arnold Ventures, and the Commonwealth Fund. The authors had no relevant disclosures.
A version of this article appeared on Medscape.com.
TOPLINE:
METHODOLOGY:
- The strength and quality of evidence supporting genome-targeted cancer drug approvals vary. A big reason is the growing number of cancer drug approvals based on surrogate endpoints, such as disease-free and progression-free survival, instead of clinical endpoints, such as overall survival or quality of life. The US Food and Drug Administration (FDA) has also approved genome-targeted cancer drugs based on phase 1 or single-arm trials.
- Given these less rigorous considerations for approval, “the validity and value of the targets and surrogate measures underlying FDA genome-targeted cancer drug approvals are uncertain,” the researchers explained.
- In the current analysis, researchers assessed the validity of the molecular targets as well as the clinical benefits of genome-targeted cancer drugs approved in the United States from 2015 to 2022 based on results from pivotal trials.
- The researchers evaluated the strength of evidence supporting molecular targetability using the European Society for Medical Oncology (ESMO) Scale for Clinical Actionability of Molecular Targets (ESCAT) and the clinical benefit using the ESMO–Magnitude of Clinical Benefit Scale (ESMO-MCBS).
- The authors defined a substantial clinical benefit as an A or B grade for curative intent and a 4 or 5 for noncurative intent. High-benefit genomic-based cancer treatments were defined as those associated with a substantial clinical benefit (ESMO-MCBS) and that qualified as ESCAT category level I-A (a clinical benefit based on prospective randomized data) or I-B (prospective nonrandomized data).
TAKEAWAY:
- The analyses focused on 50 molecular-targeted cancer drugs covering 84 indications. Of which, 45 indications (54%) were approved based on phase 1 or 2 pivotal trials, 45 (54%) were supported by single-arm pivotal trials and the remaining 39 (46%) by randomized trial, and 48 (57%) were approved based on subgroup analyses.
- Among the 84 indications, more than half (55%) of the pivotal trials supporting approval used overall response rate as a primary endpoint, 31% used progression-free survival, and 6% used disease-free survival. Only seven indications (8%) were supported by pivotal trials demonstrating an improvement in overall survival.
- Among the 84 trials, 24 (29%) met the ESMO-MCBS threshold for substantial clinical benefit.
- Overall, when combining all ratings, only 24 of the 84 indications (29%) were considered high-benefit genomic-based cancer treatments.
IN PRACTICE:
“We applied the ESMO-MCBS and ESCAT value frameworks to identify therapies and molecular targets providing high clinical value that should be widely available to patients” and “found that drug indications supported by these characteristics represent a minority of cancer drug approvals in recent years,” the authors said. Using these value frameworks could help payers, governments, and individual patients “prioritize the availability of high-value molecular-targeted therapies.”
SOURCE:
The study, with first author Ariadna Tibau, MD, PhD, Brigham and Women’s Hospital and Harvard Medical School, Boston, was published online in JAMA Oncology.
LIMITATIONS:
The study evaluated only trials that supported regulatory approval and did not include outcomes of postapproval clinical studies, which could lead to changes in ESMO-MCBS grades and ESCAT levels of evidence over time.
DISCLOSURES:
The study was funded by the Kaiser Permanente Institute for Health Policy, Arnold Ventures, and the Commonwealth Fund. The authors had no relevant disclosures.
A version of this article appeared on Medscape.com.
No Routine Cancer Screening Option? New MCED Tests May Help
Analyses presented during a session at the American Association for Cancer Research annual meeting, revealed that three new MCED tests — CanScan, MERCURY, and OncoSeek — could detect a range of cancers and recognize the tissue of origin with high accuracy. One — OncoSeek — could also provide an affordable cancer screening option for individuals living in lower-income countries.
The need for these noninvasive liquid biopsy tests that can accurately identify multiple cancer types with a single blood draw, especially cancers without routine screening strategies, is pressing. “We know that the current cancer standard of care screening will identify less than 50% of all cancers, while more than 50% of all cancer deaths occur in types of cancer with no recommended screening,” said co-moderator Marie E. Wood, MD, of the University of Colorado Anschutz Medical Campus, in Aurora, Colorado.
That being said, “the clinical utility of multicancer detection tests has not been established and we’re concerned about issues of overdiagnosis and overtreatment,” she noted.
The Early Data
One new MCED test called CanScan, developed by Geneseeq Technology, uses plasma cell-free DNA fragment patterns to detect cancer signals as well as identify the tissue of origin across 13 cancer types.
Overall, the CanScan test covers cancer types that contribute to two thirds of new cancer cases and 74% of morality globally, said presenter Shanshan Yang, of Geneseeq Research Institute, in Nanjing, China.
However, only five of these cancer types have screening recommendations issued by the US Preventive Services Task Force (USPSTF), Dr. Yang added.
The interim data comes from an ongoing large-scale prospective study evaluating the MCED test in a cohort of asymptomatic individuals between ages 45 and 75 years with an average risk for cancer and no cancer-related symptoms on enrollment.
Patients at baseline had their blood collected for the CanScan test and subsequently received annual routine physical exams once a year for 3 consecutive years, with an additional 2 years of follow-up.
The analysis included 3724 participants with analyzable samples at the data cutoff in September 2023. Among the 3724 participants, 29 had confirmed cancer diagnoses. Among these cases, 14 patients had their cancer confirmed through USPSTF recommended screening and 15 were detected through outside of standard USPSTF screening, such as a thyroid ultrasound, Dr. Yang explained.
Almost 90% of the cancers (26 of 29) were detected in the stage I or II, and eight (27.5%) were not one of the test’s 13 targeted cancer types.
The CanScan test had a sensitivity of 55.2%, identifying 16 of 29 of the patients with cancer, including 10 of 21 individuals with stage I (47.6%), and two of three with stage II (66.7%).
The test had a high specificity of 97.9%, meaning out of 100 people screened, only two had false negative findings.
Among the 15 patients who had their cancer detected outside of USPSTF screening recommendations, eight (53.3%) were found using a CanScan test, including patients with liver and endometrial cancers.
Compared with a positive predictive value of (PPV) of 1.6% with screening or physical exam methods alone, the CanScan test had a PPV of 17.4%, Dr. Yang reported.
“The MCED test holds significant potential for early cancer screening in asymptomatic populations,” Dr. Yang and colleagues concluded.
Another new MCED test called MERCURY, also developed by Geneseeq Technology and presented during the session, used a similar method to detect cancer signals and predict the tissue of origin across 13 cancer types.
The researchers initially validated the test using 3076 patients with cancer and 3477 healthy controls with a target specificity of 99%. In this group, researchers reported a sensitivity of 0.865 and a specificity of 0.989.
The team then performed an independent validation analysis with 1465 participants, 732 with cancer and 733 with no cancer, and confirmed a high sensitivity and specificity of 0.874 and 0.978, respectively. The sensitivity increased incrementally by cancer stage — 0.768 for stage I, 0.840 for stage II, 0.923 for stage III, and 0.971 for stage IV.
The test identified the tissue of origin with high accuracy, the researchers noted, but cautioned that the test needs “to be further validated in a prospective cohort study.”
MCED in Low-Income Settings
The session also featured findings on a new affordable MCED test called OncoSeek, which could provide greater access to cancer testing in low- and middle-income countries.
The OncoSeek algorithm identifies the presence of cancer using seven protein tumor markers alongside clinical information, such as gender and age. Like other tests, the test also predicts the possible tissue of origin.
The test can be run on clinical protein assay instruments that are already widely available, such as Roche cobas analyzer, Mao Mao, MD, PhD, the founder and CEO of SeekIn, of Shenzhen, China, told this news organization.
This “feature makes the test accessible worldwide, even in low- and middle-income countries,” he said. “These instruments are fully-automated and part of today’s clinical practice. Therefore, the test does not require additional infrastructure building and lab personal training.”
Another notable advantage: the OncoSeek test only costs about $20, compared with other MCED tests, which can cost anywhere from $200 to $1000.
To validate the technology in a large, diverse cohort, Dr. Mao and colleagues enrolled approximately 10,000 participants, including 2003 cancer cases and 7888 non-cancer cases.
Peripheral blood was collected from each participant and analyzed using a panel of the seven protein tumor markers — AFP, CA125, CA15-3, CA19-9, CA72-4, CEA, and CYFRA 21-1.
To reduce the risk for false positive findings, the team designed the OncoSeek algorithm to achieve a specificity of 93%. Dr. Mao and colleagues found a sensitivity of 51.7%, resulting in an overall accuracy of 84.6%.
The performance was consistent in additional validation cohorts in Brazil, China, and the United States, with sensitivities ranging from 39.0% to 77.6% for detecting nine common cancer types, including breast, colorectal, liver, lung, lymphoma, esophagus, ovary, pancreas, and stomach. The sensitivity for pancreatic cancer was at the high end of 77.6%.
The test could predict the tissue of origin in about two thirds of cases.
Given its low cost, OncoSeek represents an affordable and accessible option for cancer screening, the authors concluded.
Overall, “I think MCEDs have the potential to enhance cancer screening,” Dr. Wood told this news organization.
Still, questions remain about the optimal use of these tests, such as whether they are best for average-risk or higher risk populations, and how to integrate them into standard screening, she said.
Dr. Wood also cautioned that the studies presented in the session represent early data, and it is likely that the numbers, such as sensitivity and specificity, will change with further prospective analyses.
And ultimately, these tests should complement, not replace, standard screening. “A negative testing should not be taken as a sign to avoid standard screening,” Dr. Wood said.
Dr. Yang is an employee of Geneseeq Technology, Inc., and Dr. Mao is an employee of SeekIn. Dr. Wood had no disclosures to report.
A version of this article appeared on Medscape.com.
Analyses presented during a session at the American Association for Cancer Research annual meeting, revealed that three new MCED tests — CanScan, MERCURY, and OncoSeek — could detect a range of cancers and recognize the tissue of origin with high accuracy. One — OncoSeek — could also provide an affordable cancer screening option for individuals living in lower-income countries.
The need for these noninvasive liquid biopsy tests that can accurately identify multiple cancer types with a single blood draw, especially cancers without routine screening strategies, is pressing. “We know that the current cancer standard of care screening will identify less than 50% of all cancers, while more than 50% of all cancer deaths occur in types of cancer with no recommended screening,” said co-moderator Marie E. Wood, MD, of the University of Colorado Anschutz Medical Campus, in Aurora, Colorado.
That being said, “the clinical utility of multicancer detection tests has not been established and we’re concerned about issues of overdiagnosis and overtreatment,” she noted.
The Early Data
One new MCED test called CanScan, developed by Geneseeq Technology, uses plasma cell-free DNA fragment patterns to detect cancer signals as well as identify the tissue of origin across 13 cancer types.
Overall, the CanScan test covers cancer types that contribute to two thirds of new cancer cases and 74% of morality globally, said presenter Shanshan Yang, of Geneseeq Research Institute, in Nanjing, China.
However, only five of these cancer types have screening recommendations issued by the US Preventive Services Task Force (USPSTF), Dr. Yang added.
The interim data comes from an ongoing large-scale prospective study evaluating the MCED test in a cohort of asymptomatic individuals between ages 45 and 75 years with an average risk for cancer and no cancer-related symptoms on enrollment.
Patients at baseline had their blood collected for the CanScan test and subsequently received annual routine physical exams once a year for 3 consecutive years, with an additional 2 years of follow-up.
The analysis included 3724 participants with analyzable samples at the data cutoff in September 2023. Among the 3724 participants, 29 had confirmed cancer diagnoses. Among these cases, 14 patients had their cancer confirmed through USPSTF recommended screening and 15 were detected through outside of standard USPSTF screening, such as a thyroid ultrasound, Dr. Yang explained.
Almost 90% of the cancers (26 of 29) were detected in the stage I or II, and eight (27.5%) were not one of the test’s 13 targeted cancer types.
The CanScan test had a sensitivity of 55.2%, identifying 16 of 29 of the patients with cancer, including 10 of 21 individuals with stage I (47.6%), and two of three with stage II (66.7%).
The test had a high specificity of 97.9%, meaning out of 100 people screened, only two had false negative findings.
Among the 15 patients who had their cancer detected outside of USPSTF screening recommendations, eight (53.3%) were found using a CanScan test, including patients with liver and endometrial cancers.
Compared with a positive predictive value of (PPV) of 1.6% with screening or physical exam methods alone, the CanScan test had a PPV of 17.4%, Dr. Yang reported.
“The MCED test holds significant potential for early cancer screening in asymptomatic populations,” Dr. Yang and colleagues concluded.
Another new MCED test called MERCURY, also developed by Geneseeq Technology and presented during the session, used a similar method to detect cancer signals and predict the tissue of origin across 13 cancer types.
The researchers initially validated the test using 3076 patients with cancer and 3477 healthy controls with a target specificity of 99%. In this group, researchers reported a sensitivity of 0.865 and a specificity of 0.989.
The team then performed an independent validation analysis with 1465 participants, 732 with cancer and 733 with no cancer, and confirmed a high sensitivity and specificity of 0.874 and 0.978, respectively. The sensitivity increased incrementally by cancer stage — 0.768 for stage I, 0.840 for stage II, 0.923 for stage III, and 0.971 for stage IV.
The test identified the tissue of origin with high accuracy, the researchers noted, but cautioned that the test needs “to be further validated in a prospective cohort study.”
MCED in Low-Income Settings
The session also featured findings on a new affordable MCED test called OncoSeek, which could provide greater access to cancer testing in low- and middle-income countries.
The OncoSeek algorithm identifies the presence of cancer using seven protein tumor markers alongside clinical information, such as gender and age. Like other tests, the test also predicts the possible tissue of origin.
The test can be run on clinical protein assay instruments that are already widely available, such as Roche cobas analyzer, Mao Mao, MD, PhD, the founder and CEO of SeekIn, of Shenzhen, China, told this news organization.
This “feature makes the test accessible worldwide, even in low- and middle-income countries,” he said. “These instruments are fully-automated and part of today’s clinical practice. Therefore, the test does not require additional infrastructure building and lab personal training.”
Another notable advantage: the OncoSeek test only costs about $20, compared with other MCED tests, which can cost anywhere from $200 to $1000.
To validate the technology in a large, diverse cohort, Dr. Mao and colleagues enrolled approximately 10,000 participants, including 2003 cancer cases and 7888 non-cancer cases.
Peripheral blood was collected from each participant and analyzed using a panel of the seven protein tumor markers — AFP, CA125, CA15-3, CA19-9, CA72-4, CEA, and CYFRA 21-1.
To reduce the risk for false positive findings, the team designed the OncoSeek algorithm to achieve a specificity of 93%. Dr. Mao and colleagues found a sensitivity of 51.7%, resulting in an overall accuracy of 84.6%.
The performance was consistent in additional validation cohorts in Brazil, China, and the United States, with sensitivities ranging from 39.0% to 77.6% for detecting nine common cancer types, including breast, colorectal, liver, lung, lymphoma, esophagus, ovary, pancreas, and stomach. The sensitivity for pancreatic cancer was at the high end of 77.6%.
The test could predict the tissue of origin in about two thirds of cases.
Given its low cost, OncoSeek represents an affordable and accessible option for cancer screening, the authors concluded.
Overall, “I think MCEDs have the potential to enhance cancer screening,” Dr. Wood told this news organization.
Still, questions remain about the optimal use of these tests, such as whether they are best for average-risk or higher risk populations, and how to integrate them into standard screening, she said.
Dr. Wood also cautioned that the studies presented in the session represent early data, and it is likely that the numbers, such as sensitivity and specificity, will change with further prospective analyses.
And ultimately, these tests should complement, not replace, standard screening. “A negative testing should not be taken as a sign to avoid standard screening,” Dr. Wood said.
Dr. Yang is an employee of Geneseeq Technology, Inc., and Dr. Mao is an employee of SeekIn. Dr. Wood had no disclosures to report.
A version of this article appeared on Medscape.com.
Analyses presented during a session at the American Association for Cancer Research annual meeting, revealed that three new MCED tests — CanScan, MERCURY, and OncoSeek — could detect a range of cancers and recognize the tissue of origin with high accuracy. One — OncoSeek — could also provide an affordable cancer screening option for individuals living in lower-income countries.
The need for these noninvasive liquid biopsy tests that can accurately identify multiple cancer types with a single blood draw, especially cancers without routine screening strategies, is pressing. “We know that the current cancer standard of care screening will identify less than 50% of all cancers, while more than 50% of all cancer deaths occur in types of cancer with no recommended screening,” said co-moderator Marie E. Wood, MD, of the University of Colorado Anschutz Medical Campus, in Aurora, Colorado.
That being said, “the clinical utility of multicancer detection tests has not been established and we’re concerned about issues of overdiagnosis and overtreatment,” she noted.
The Early Data
One new MCED test called CanScan, developed by Geneseeq Technology, uses plasma cell-free DNA fragment patterns to detect cancer signals as well as identify the tissue of origin across 13 cancer types.
Overall, the CanScan test covers cancer types that contribute to two thirds of new cancer cases and 74% of morality globally, said presenter Shanshan Yang, of Geneseeq Research Institute, in Nanjing, China.
However, only five of these cancer types have screening recommendations issued by the US Preventive Services Task Force (USPSTF), Dr. Yang added.
The interim data comes from an ongoing large-scale prospective study evaluating the MCED test in a cohort of asymptomatic individuals between ages 45 and 75 years with an average risk for cancer and no cancer-related symptoms on enrollment.
Patients at baseline had their blood collected for the CanScan test and subsequently received annual routine physical exams once a year for 3 consecutive years, with an additional 2 years of follow-up.
The analysis included 3724 participants with analyzable samples at the data cutoff in September 2023. Among the 3724 participants, 29 had confirmed cancer diagnoses. Among these cases, 14 patients had their cancer confirmed through USPSTF recommended screening and 15 were detected through outside of standard USPSTF screening, such as a thyroid ultrasound, Dr. Yang explained.
Almost 90% of the cancers (26 of 29) were detected in the stage I or II, and eight (27.5%) were not one of the test’s 13 targeted cancer types.
The CanScan test had a sensitivity of 55.2%, identifying 16 of 29 of the patients with cancer, including 10 of 21 individuals with stage I (47.6%), and two of three with stage II (66.7%).
The test had a high specificity of 97.9%, meaning out of 100 people screened, only two had false negative findings.
Among the 15 patients who had their cancer detected outside of USPSTF screening recommendations, eight (53.3%) were found using a CanScan test, including patients with liver and endometrial cancers.
Compared with a positive predictive value of (PPV) of 1.6% with screening or physical exam methods alone, the CanScan test had a PPV of 17.4%, Dr. Yang reported.
“The MCED test holds significant potential for early cancer screening in asymptomatic populations,” Dr. Yang and colleagues concluded.
Another new MCED test called MERCURY, also developed by Geneseeq Technology and presented during the session, used a similar method to detect cancer signals and predict the tissue of origin across 13 cancer types.
The researchers initially validated the test using 3076 patients with cancer and 3477 healthy controls with a target specificity of 99%. In this group, researchers reported a sensitivity of 0.865 and a specificity of 0.989.
The team then performed an independent validation analysis with 1465 participants, 732 with cancer and 733 with no cancer, and confirmed a high sensitivity and specificity of 0.874 and 0.978, respectively. The sensitivity increased incrementally by cancer stage — 0.768 for stage I, 0.840 for stage II, 0.923 for stage III, and 0.971 for stage IV.
The test identified the tissue of origin with high accuracy, the researchers noted, but cautioned that the test needs “to be further validated in a prospective cohort study.”
MCED in Low-Income Settings
The session also featured findings on a new affordable MCED test called OncoSeek, which could provide greater access to cancer testing in low- and middle-income countries.
The OncoSeek algorithm identifies the presence of cancer using seven protein tumor markers alongside clinical information, such as gender and age. Like other tests, the test also predicts the possible tissue of origin.
The test can be run on clinical protein assay instruments that are already widely available, such as Roche cobas analyzer, Mao Mao, MD, PhD, the founder and CEO of SeekIn, of Shenzhen, China, told this news organization.
This “feature makes the test accessible worldwide, even in low- and middle-income countries,” he said. “These instruments are fully-automated and part of today’s clinical practice. Therefore, the test does not require additional infrastructure building and lab personal training.”
Another notable advantage: the OncoSeek test only costs about $20, compared with other MCED tests, which can cost anywhere from $200 to $1000.
To validate the technology in a large, diverse cohort, Dr. Mao and colleagues enrolled approximately 10,000 participants, including 2003 cancer cases and 7888 non-cancer cases.
Peripheral blood was collected from each participant and analyzed using a panel of the seven protein tumor markers — AFP, CA125, CA15-3, CA19-9, CA72-4, CEA, and CYFRA 21-1.
To reduce the risk for false positive findings, the team designed the OncoSeek algorithm to achieve a specificity of 93%. Dr. Mao and colleagues found a sensitivity of 51.7%, resulting in an overall accuracy of 84.6%.
The performance was consistent in additional validation cohorts in Brazil, China, and the United States, with sensitivities ranging from 39.0% to 77.6% for detecting nine common cancer types, including breast, colorectal, liver, lung, lymphoma, esophagus, ovary, pancreas, and stomach. The sensitivity for pancreatic cancer was at the high end of 77.6%.
The test could predict the tissue of origin in about two thirds of cases.
Given its low cost, OncoSeek represents an affordable and accessible option for cancer screening, the authors concluded.
Overall, “I think MCEDs have the potential to enhance cancer screening,” Dr. Wood told this news organization.
Still, questions remain about the optimal use of these tests, such as whether they are best for average-risk or higher risk populations, and how to integrate them into standard screening, she said.
Dr. Wood also cautioned that the studies presented in the session represent early data, and it is likely that the numbers, such as sensitivity and specificity, will change with further prospective analyses.
And ultimately, these tests should complement, not replace, standard screening. “A negative testing should not be taken as a sign to avoid standard screening,” Dr. Wood said.
Dr. Yang is an employee of Geneseeq Technology, Inc., and Dr. Mao is an employee of SeekIn. Dr. Wood had no disclosures to report.
A version of this article appeared on Medscape.com.
Oncologists Voice Ethical Concerns Over AI in Cancer Care
TOPLINE:
Most respondents, for instance, said patients should not be expected to understand how AI tools work, but many also felt patients could make treatment decisions based on AI-generated recommendations. Most oncologists also felt responsible for protecting patients from biased AI, but few were confident that they could do so.
METHODOLOGY:
- The US Food and Drug Administration (FDA) has for use in various medical specialties over the past few decades, and increasingly, AI tools are being integrated into cancer care.
- However, the uptake of these tools in oncology has raised ethical questions and concerns, including challenges with AI bias, error, or misuse, as well as issues explaining how an AI model reached a result.
- In the current study, researchers asked 204 oncologists from 37 states for their views on the ethical implications of using AI for cancer care.
- Among the survey respondents, 64% were men and 63% were non-Hispanic White; 29% were from academic practices, 47% had received some education on AI use in healthcare, and 45% were familiar with clinical decision models.
- The researchers assessed respondents’ answers to various questions, including whether to provide informed consent for AI use and how oncologists would approach a scenario where the AI model and the oncologist recommended a different treatment regimen.
TAKEAWAY:
- Overall, 81% of oncologists supported having patient consent to use an AI model during treatment decisions, and 85% felt that oncologists needed to be able to explain an AI-based clinical decision model to use it in the clinic; however, only 23% felt that patients also needed to be able to explain an AI model.
- When an AI decision model recommended a different treatment regimen than the treating oncologist, the most common response (36.8%) was to present both options to the patient and let the patient decide. Oncologists from academic settings were about 2.5 times more likely than those from other settings to let the patient decide. About 34% of respondents said they would present both options but recommend the oncologist’s regimen, whereas about 22% said they would present both but recommend the AI’s regimen. A small percentage would only present the oncologist’s regimen (5%) or the AI’s regimen (about 2.5%).
- About three of four respondents (76.5%) agreed that oncologists should protect patients from biased AI tools; however, only about one of four (27.9%) felt confident they could identify biased AI models.
- Most oncologists (91%) felt that AI developers were responsible for the medico-legal problems associated with AI use; less than half (47%) said oncologists or hospitals (43%) shared this responsibility.
IN PRACTICE:
“Together, these data characterize barriers that may impede the ethical adoption of AI into cancer care. The findings suggest that the implementation of AI in oncology must include rigorous assessments of its effect on care decisions, as well as decisional responsibility when problems related to AI use arise,” the authors concluded.
SOURCE:
The study, with first author Andrew Hantel, MD, from Dana-Farber Cancer Institute, Boston, was published last month in JAMA Network Open.
LIMITATIONS:
The study had a moderate sample size and response rate, although demographics of participating oncologists appear to be nationally representative. The cross-sectional study design limited the generalizability of the findings over time as AI is integrated into cancer care.
DISCLOSURES:
The study was funded by the National Cancer Institute, the Dana-Farber McGraw/Patterson Research Fund, and the Mark Foundation Emerging Leader Award. Dr. Hantel reported receiving personal fees from AbbVie, AstraZeneca, the American Journal of Managed Care, Genentech, and GSK.
A version of this article appeared on Medscape.com.
TOPLINE:
Most respondents, for instance, said patients should not be expected to understand how AI tools work, but many also felt patients could make treatment decisions based on AI-generated recommendations. Most oncologists also felt responsible for protecting patients from biased AI, but few were confident that they could do so.
METHODOLOGY:
- The US Food and Drug Administration (FDA) has for use in various medical specialties over the past few decades, and increasingly, AI tools are being integrated into cancer care.
- However, the uptake of these tools in oncology has raised ethical questions and concerns, including challenges with AI bias, error, or misuse, as well as issues explaining how an AI model reached a result.
- In the current study, researchers asked 204 oncologists from 37 states for their views on the ethical implications of using AI for cancer care.
- Among the survey respondents, 64% were men and 63% were non-Hispanic White; 29% were from academic practices, 47% had received some education on AI use in healthcare, and 45% were familiar with clinical decision models.
- The researchers assessed respondents’ answers to various questions, including whether to provide informed consent for AI use and how oncologists would approach a scenario where the AI model and the oncologist recommended a different treatment regimen.
TAKEAWAY:
- Overall, 81% of oncologists supported having patient consent to use an AI model during treatment decisions, and 85% felt that oncologists needed to be able to explain an AI-based clinical decision model to use it in the clinic; however, only 23% felt that patients also needed to be able to explain an AI model.
- When an AI decision model recommended a different treatment regimen than the treating oncologist, the most common response (36.8%) was to present both options to the patient and let the patient decide. Oncologists from academic settings were about 2.5 times more likely than those from other settings to let the patient decide. About 34% of respondents said they would present both options but recommend the oncologist’s regimen, whereas about 22% said they would present both but recommend the AI’s regimen. A small percentage would only present the oncologist’s regimen (5%) or the AI’s regimen (about 2.5%).
- About three of four respondents (76.5%) agreed that oncologists should protect patients from biased AI tools; however, only about one of four (27.9%) felt confident they could identify biased AI models.
- Most oncologists (91%) felt that AI developers were responsible for the medico-legal problems associated with AI use; less than half (47%) said oncologists or hospitals (43%) shared this responsibility.
IN PRACTICE:
“Together, these data characterize barriers that may impede the ethical adoption of AI into cancer care. The findings suggest that the implementation of AI in oncology must include rigorous assessments of its effect on care decisions, as well as decisional responsibility when problems related to AI use arise,” the authors concluded.
SOURCE:
The study, with first author Andrew Hantel, MD, from Dana-Farber Cancer Institute, Boston, was published last month in JAMA Network Open.
LIMITATIONS:
The study had a moderate sample size and response rate, although demographics of participating oncologists appear to be nationally representative. The cross-sectional study design limited the generalizability of the findings over time as AI is integrated into cancer care.
DISCLOSURES:
The study was funded by the National Cancer Institute, the Dana-Farber McGraw/Patterson Research Fund, and the Mark Foundation Emerging Leader Award. Dr. Hantel reported receiving personal fees from AbbVie, AstraZeneca, the American Journal of Managed Care, Genentech, and GSK.
A version of this article appeared on Medscape.com.
TOPLINE:
Most respondents, for instance, said patients should not be expected to understand how AI tools work, but many also felt patients could make treatment decisions based on AI-generated recommendations. Most oncologists also felt responsible for protecting patients from biased AI, but few were confident that they could do so.
METHODOLOGY:
- The US Food and Drug Administration (FDA) has for use in various medical specialties over the past few decades, and increasingly, AI tools are being integrated into cancer care.
- However, the uptake of these tools in oncology has raised ethical questions and concerns, including challenges with AI bias, error, or misuse, as well as issues explaining how an AI model reached a result.
- In the current study, researchers asked 204 oncologists from 37 states for their views on the ethical implications of using AI for cancer care.
- Among the survey respondents, 64% were men and 63% were non-Hispanic White; 29% were from academic practices, 47% had received some education on AI use in healthcare, and 45% were familiar with clinical decision models.
- The researchers assessed respondents’ answers to various questions, including whether to provide informed consent for AI use and how oncologists would approach a scenario where the AI model and the oncologist recommended a different treatment regimen.
TAKEAWAY:
- Overall, 81% of oncologists supported having patient consent to use an AI model during treatment decisions, and 85% felt that oncologists needed to be able to explain an AI-based clinical decision model to use it in the clinic; however, only 23% felt that patients also needed to be able to explain an AI model.
- When an AI decision model recommended a different treatment regimen than the treating oncologist, the most common response (36.8%) was to present both options to the patient and let the patient decide. Oncologists from academic settings were about 2.5 times more likely than those from other settings to let the patient decide. About 34% of respondents said they would present both options but recommend the oncologist’s regimen, whereas about 22% said they would present both but recommend the AI’s regimen. A small percentage would only present the oncologist’s regimen (5%) or the AI’s regimen (about 2.5%).
- About three of four respondents (76.5%) agreed that oncologists should protect patients from biased AI tools; however, only about one of four (27.9%) felt confident they could identify biased AI models.
- Most oncologists (91%) felt that AI developers were responsible for the medico-legal problems associated with AI use; less than half (47%) said oncologists or hospitals (43%) shared this responsibility.
IN PRACTICE:
“Together, these data characterize barriers that may impede the ethical adoption of AI into cancer care. The findings suggest that the implementation of AI in oncology must include rigorous assessments of its effect on care decisions, as well as decisional responsibility when problems related to AI use arise,” the authors concluded.
SOURCE:
The study, with first author Andrew Hantel, MD, from Dana-Farber Cancer Institute, Boston, was published last month in JAMA Network Open.
LIMITATIONS:
The study had a moderate sample size and response rate, although demographics of participating oncologists appear to be nationally representative. The cross-sectional study design limited the generalizability of the findings over time as AI is integrated into cancer care.
DISCLOSURES:
The study was funded by the National Cancer Institute, the Dana-Farber McGraw/Patterson Research Fund, and the Mark Foundation Emerging Leader Award. Dr. Hantel reported receiving personal fees from AbbVie, AstraZeneca, the American Journal of Managed Care, Genentech, and GSK.
A version of this article appeared on Medscape.com.
Consider Skin Cancer, Infection Risks in Solid Organ Transplant Recipients
SAN DIEGO — because of their suppressed immune systems.
“There are over 450,000 people with a solid organ transplant living in the United States. If you do the math, that works out to about 40 organ transplant recipients for every dermatologist, so there’s a lot of them out there for us to take care of,” Sean Christensen, MD, PhD, associate professor of dermatology, Yale University, New Haven, Connecticut, said at the annual meeting of the American Academy of Dermatology (AAD). “If we expand that umbrella to include all types of immunosuppression, that’s over 4 million adults in the US.”
Dr. Christensen encouraged dermatologists to be aware of the varying risks for immunosuppressive drugs and best screening practices for these patients, and to take advantage of a validated skin cancer risk assessment tool for transplant patients.
During his presentation, he highlighted five classes of immunosuppressive drugs and their associated skin cancer risks:
- Calcineurin inhibitors (tacrolimus or cyclosporine), which cause severe immune suppression and pose a severe skin cancer risk. They may also cause gingival hyperplasia and sebaceous hyperplasia.
- Antimetabolites (mycophenolate mofetil or azathioprine), which cause moderate to severe immune suppression and pose a severe skin cancer risk.
- Mammalian target of rapamycin inhibitors (sirolimus or everolimus), which cause severe immune suppression and pose a moderate skin cancer risk. They also impair wound healing.
- Corticosteroids (prednisone), which cause mild to severe immune suppression and pose a minimal skin cancer risk.
- A decoy receptor protein (belatacept), which causes severe immune suppression and poses a mild skin cancer risk.
“Most of our solid-organ transplant recipients will be on both a calcineurin inhibitor and an antimetabolite,” Dr. Christensen said. “In addition to the skin cancer risk associated with immunosuppression, there is an additive risk” that is a direct effect of these medications on the skin. “That means our transplant recipients have a severely and disproportionate increase in skin cancer,” he noted.
Up to half of solid-organ transplant recipients will develop skin cancer, Dr. Christensen said. These patients have a sixfold to 10-fold increased risk for basal cell carcinoma (BCC), a 35- to 65-fold increased risk for squamous cell carcinoma (SCC), a twofold to sevenfold increased risk for melanoma, and a 16- to 100-fold increased risk for Merkel cell carcinoma.
Transplant recipients with SCC, he said, have a twofold to threefold higher risk for metastasis (4%-8% nodal metastasis) and twofold to fivefold higher risk for death (2%-7% mortality) from SCC.
As for other kinds of immunosuppression, HIV positivity, treatment with 6-mercaptopurine or azathioprine (for inflammatory bowel disease and rheumatoid arthritis), and antitumor necrosis factor agents (for psoriasis, inflammatory bowel disease, and rheumatoid arthritis) have been linked in studies to a higher risk for nonmelanoma skin cancer.
Dr. Christensen also highlighted graft-versus-host disease (GVHD). “It does look like there is a disproportionate and increased risk of SCC of the oropharynx and of the skin in patients who have chronic GVHD. This is probably due to a combination of both the immunosuppressive medications that are required but also from chronic and ongoing inflammation in the skin.”
Chronic GVHD has been linked to a 5.3-fold increase in the risk for SCC and a twofold increase in the risk for BCC, he added.
Moreover, new medications for treating GVHD have been linked to an increased risk for SCC, including a 3.2-fold increased risk for SCC associated with ruxolitinib, a Janus kinase (JAK) 1 and JAK2 inhibitor, in a study of patients with polycythemia vera and myelofibrosis; and a case report of SCC in a patient treated with belumosudil, a rho-associated coiled-coil-containing protein kinase-2 kinase inhibitor, for chronic GVHD. Risk for SCC appears to increase based on duration of use with voriconazole, an antifungal, which, he said, is a potent photosynthesizer.
Dr. Christensen also noted the higher risk for infections in immunocompromised patients and added that these patients can develop inflammatory disease despite immunosuppression:
Staphylococcus, Streptococcus, and Dermatophytes are the most common skin pathogens in these patients. There’s a significantly increased risk for reactivation of herpes simplex, varicella-zoster viruses, and cytomegalovirus. Opportunistic and disseminated fungal infections, such as mycobacteria, Candida, histoplasma, cryptococcus, aspergillus, and mucormycosis, can also appear.
More than 80% of transplant recipients develop molluscum and verruca vulgaris/human papillomavirus infection. They may also develop noninfectious inflammatory dermatoses.
Risk Calculator
What can dermatologists do to help transplant patients? Dr. Christensen highlighted the Skin and UV Neoplasia Transplant Risk Assessment Calculator, which predicts skin cancer risk based on points given for race, gender, skin cancer history, age at transplant, and site of transplant.
The tool, validated in a 2023 study of transplant recipients in Europe, is available online and as an app. It makes recommendations to users about when patients should have initial skin screening exams. Those with the most risk — 45% at 5 years — should be screened within 6 months. “We can use [the tool] to triage these cases when we first meet them and get them plugged into the appropriate care,” Dr. Christensen said.
He recommended seeing high-risk patients at least annually. Patients with a prior SCC and a heavy burden of actinic keratosis should be followed more frequently, he said.
In regard to SCC, he highlighted a 2024 study of solid organ transplant recipients that found the risk for a second SCC after a first SCC was 74%, the risk for a third SCC after a second SCC was 83%, and the risk for another SCC after five SCCs was 92%.
Dr. Christensen disclosed relationships with Canfield Scientific Inc. (consulting), Inhibitor Therapeutics (advisory board), and Sol-Gel Technologies Ltd. (grants/research funding).
A version of this article first appeared on Medscape.com.
SAN DIEGO — because of their suppressed immune systems.
“There are over 450,000 people with a solid organ transplant living in the United States. If you do the math, that works out to about 40 organ transplant recipients for every dermatologist, so there’s a lot of them out there for us to take care of,” Sean Christensen, MD, PhD, associate professor of dermatology, Yale University, New Haven, Connecticut, said at the annual meeting of the American Academy of Dermatology (AAD). “If we expand that umbrella to include all types of immunosuppression, that’s over 4 million adults in the US.”
Dr. Christensen encouraged dermatologists to be aware of the varying risks for immunosuppressive drugs and best screening practices for these patients, and to take advantage of a validated skin cancer risk assessment tool for transplant patients.
During his presentation, he highlighted five classes of immunosuppressive drugs and their associated skin cancer risks:
- Calcineurin inhibitors (tacrolimus or cyclosporine), which cause severe immune suppression and pose a severe skin cancer risk. They may also cause gingival hyperplasia and sebaceous hyperplasia.
- Antimetabolites (mycophenolate mofetil or azathioprine), which cause moderate to severe immune suppression and pose a severe skin cancer risk.
- Mammalian target of rapamycin inhibitors (sirolimus or everolimus), which cause severe immune suppression and pose a moderate skin cancer risk. They also impair wound healing.
- Corticosteroids (prednisone), which cause mild to severe immune suppression and pose a minimal skin cancer risk.
- A decoy receptor protein (belatacept), which causes severe immune suppression and poses a mild skin cancer risk.
“Most of our solid-organ transplant recipients will be on both a calcineurin inhibitor and an antimetabolite,” Dr. Christensen said. “In addition to the skin cancer risk associated with immunosuppression, there is an additive risk” that is a direct effect of these medications on the skin. “That means our transplant recipients have a severely and disproportionate increase in skin cancer,” he noted.
Up to half of solid-organ transplant recipients will develop skin cancer, Dr. Christensen said. These patients have a sixfold to 10-fold increased risk for basal cell carcinoma (BCC), a 35- to 65-fold increased risk for squamous cell carcinoma (SCC), a twofold to sevenfold increased risk for melanoma, and a 16- to 100-fold increased risk for Merkel cell carcinoma.
Transplant recipients with SCC, he said, have a twofold to threefold higher risk for metastasis (4%-8% nodal metastasis) and twofold to fivefold higher risk for death (2%-7% mortality) from SCC.
As for other kinds of immunosuppression, HIV positivity, treatment with 6-mercaptopurine or azathioprine (for inflammatory bowel disease and rheumatoid arthritis), and antitumor necrosis factor agents (for psoriasis, inflammatory bowel disease, and rheumatoid arthritis) have been linked in studies to a higher risk for nonmelanoma skin cancer.
Dr. Christensen also highlighted graft-versus-host disease (GVHD). “It does look like there is a disproportionate and increased risk of SCC of the oropharynx and of the skin in patients who have chronic GVHD. This is probably due to a combination of both the immunosuppressive medications that are required but also from chronic and ongoing inflammation in the skin.”
Chronic GVHD has been linked to a 5.3-fold increase in the risk for SCC and a twofold increase in the risk for BCC, he added.
Moreover, new medications for treating GVHD have been linked to an increased risk for SCC, including a 3.2-fold increased risk for SCC associated with ruxolitinib, a Janus kinase (JAK) 1 and JAK2 inhibitor, in a study of patients with polycythemia vera and myelofibrosis; and a case report of SCC in a patient treated with belumosudil, a rho-associated coiled-coil-containing protein kinase-2 kinase inhibitor, for chronic GVHD. Risk for SCC appears to increase based on duration of use with voriconazole, an antifungal, which, he said, is a potent photosynthesizer.
Dr. Christensen also noted the higher risk for infections in immunocompromised patients and added that these patients can develop inflammatory disease despite immunosuppression:
Staphylococcus, Streptococcus, and Dermatophytes are the most common skin pathogens in these patients. There’s a significantly increased risk for reactivation of herpes simplex, varicella-zoster viruses, and cytomegalovirus. Opportunistic and disseminated fungal infections, such as mycobacteria, Candida, histoplasma, cryptococcus, aspergillus, and mucormycosis, can also appear.
More than 80% of transplant recipients develop molluscum and verruca vulgaris/human papillomavirus infection. They may also develop noninfectious inflammatory dermatoses.
Risk Calculator
What can dermatologists do to help transplant patients? Dr. Christensen highlighted the Skin and UV Neoplasia Transplant Risk Assessment Calculator, which predicts skin cancer risk based on points given for race, gender, skin cancer history, age at transplant, and site of transplant.
The tool, validated in a 2023 study of transplant recipients in Europe, is available online and as an app. It makes recommendations to users about when patients should have initial skin screening exams. Those with the most risk — 45% at 5 years — should be screened within 6 months. “We can use [the tool] to triage these cases when we first meet them and get them plugged into the appropriate care,” Dr. Christensen said.
He recommended seeing high-risk patients at least annually. Patients with a prior SCC and a heavy burden of actinic keratosis should be followed more frequently, he said.
In regard to SCC, he highlighted a 2024 study of solid organ transplant recipients that found the risk for a second SCC after a first SCC was 74%, the risk for a third SCC after a second SCC was 83%, and the risk for another SCC after five SCCs was 92%.
Dr. Christensen disclosed relationships with Canfield Scientific Inc. (consulting), Inhibitor Therapeutics (advisory board), and Sol-Gel Technologies Ltd. (grants/research funding).
A version of this article first appeared on Medscape.com.
SAN DIEGO — because of their suppressed immune systems.
“There are over 450,000 people with a solid organ transplant living in the United States. If you do the math, that works out to about 40 organ transplant recipients for every dermatologist, so there’s a lot of them out there for us to take care of,” Sean Christensen, MD, PhD, associate professor of dermatology, Yale University, New Haven, Connecticut, said at the annual meeting of the American Academy of Dermatology (AAD). “If we expand that umbrella to include all types of immunosuppression, that’s over 4 million adults in the US.”
Dr. Christensen encouraged dermatologists to be aware of the varying risks for immunosuppressive drugs and best screening practices for these patients, and to take advantage of a validated skin cancer risk assessment tool for transplant patients.
During his presentation, he highlighted five classes of immunosuppressive drugs and their associated skin cancer risks:
- Calcineurin inhibitors (tacrolimus or cyclosporine), which cause severe immune suppression and pose a severe skin cancer risk. They may also cause gingival hyperplasia and sebaceous hyperplasia.
- Antimetabolites (mycophenolate mofetil or azathioprine), which cause moderate to severe immune suppression and pose a severe skin cancer risk.
- Mammalian target of rapamycin inhibitors (sirolimus or everolimus), which cause severe immune suppression and pose a moderate skin cancer risk. They also impair wound healing.
- Corticosteroids (prednisone), which cause mild to severe immune suppression and pose a minimal skin cancer risk.
- A decoy receptor protein (belatacept), which causes severe immune suppression and poses a mild skin cancer risk.
“Most of our solid-organ transplant recipients will be on both a calcineurin inhibitor and an antimetabolite,” Dr. Christensen said. “In addition to the skin cancer risk associated with immunosuppression, there is an additive risk” that is a direct effect of these medications on the skin. “That means our transplant recipients have a severely and disproportionate increase in skin cancer,” he noted.
Up to half of solid-organ transplant recipients will develop skin cancer, Dr. Christensen said. These patients have a sixfold to 10-fold increased risk for basal cell carcinoma (BCC), a 35- to 65-fold increased risk for squamous cell carcinoma (SCC), a twofold to sevenfold increased risk for melanoma, and a 16- to 100-fold increased risk for Merkel cell carcinoma.
Transplant recipients with SCC, he said, have a twofold to threefold higher risk for metastasis (4%-8% nodal metastasis) and twofold to fivefold higher risk for death (2%-7% mortality) from SCC.
As for other kinds of immunosuppression, HIV positivity, treatment with 6-mercaptopurine or azathioprine (for inflammatory bowel disease and rheumatoid arthritis), and antitumor necrosis factor agents (for psoriasis, inflammatory bowel disease, and rheumatoid arthritis) have been linked in studies to a higher risk for nonmelanoma skin cancer.
Dr. Christensen also highlighted graft-versus-host disease (GVHD). “It does look like there is a disproportionate and increased risk of SCC of the oropharynx and of the skin in patients who have chronic GVHD. This is probably due to a combination of both the immunosuppressive medications that are required but also from chronic and ongoing inflammation in the skin.”
Chronic GVHD has been linked to a 5.3-fold increase in the risk for SCC and a twofold increase in the risk for BCC, he added.
Moreover, new medications for treating GVHD have been linked to an increased risk for SCC, including a 3.2-fold increased risk for SCC associated with ruxolitinib, a Janus kinase (JAK) 1 and JAK2 inhibitor, in a study of patients with polycythemia vera and myelofibrosis; and a case report of SCC in a patient treated with belumosudil, a rho-associated coiled-coil-containing protein kinase-2 kinase inhibitor, for chronic GVHD. Risk for SCC appears to increase based on duration of use with voriconazole, an antifungal, which, he said, is a potent photosynthesizer.
Dr. Christensen also noted the higher risk for infections in immunocompromised patients and added that these patients can develop inflammatory disease despite immunosuppression:
Staphylococcus, Streptococcus, and Dermatophytes are the most common skin pathogens in these patients. There’s a significantly increased risk for reactivation of herpes simplex, varicella-zoster viruses, and cytomegalovirus. Opportunistic and disseminated fungal infections, such as mycobacteria, Candida, histoplasma, cryptococcus, aspergillus, and mucormycosis, can also appear.
More than 80% of transplant recipients develop molluscum and verruca vulgaris/human papillomavirus infection. They may also develop noninfectious inflammatory dermatoses.
Risk Calculator
What can dermatologists do to help transplant patients? Dr. Christensen highlighted the Skin and UV Neoplasia Transplant Risk Assessment Calculator, which predicts skin cancer risk based on points given for race, gender, skin cancer history, age at transplant, and site of transplant.
The tool, validated in a 2023 study of transplant recipients in Europe, is available online and as an app. It makes recommendations to users about when patients should have initial skin screening exams. Those with the most risk — 45% at 5 years — should be screened within 6 months. “We can use [the tool] to triage these cases when we first meet them and get them plugged into the appropriate care,” Dr. Christensen said.
He recommended seeing high-risk patients at least annually. Patients with a prior SCC and a heavy burden of actinic keratosis should be followed more frequently, he said.
In regard to SCC, he highlighted a 2024 study of solid organ transplant recipients that found the risk for a second SCC after a first SCC was 74%, the risk for a third SCC after a second SCC was 83%, and the risk for another SCC after five SCCs was 92%.
Dr. Christensen disclosed relationships with Canfield Scientific Inc. (consulting), Inhibitor Therapeutics (advisory board), and Sol-Gel Technologies Ltd. (grants/research funding).
A version of this article first appeared on Medscape.com.
FROM AAD 2024
Circulating Tumor DNA Predicts Early Treatment Response in Patients With HER2-Positive Cancers
This was the main finding of new data presented by study author Razelle Kurzrock, MD, at the American Association for Cancer Research annual meeting.
“We found that on-treatment ctDNA can detect progression before standard-of-care response assessments. These data suggest that monitoring ctDNA can provide clinicians with important prognostic information that may guide treatment decisions,” Dr. Kurzrock, professor at the Medical College of Wisconsin, Milwaukee, said during her presentation.
Commenting on the clinical implications of these findings during an interview, she said the results suggest that ctDNA dynamics provide an early window into predicting response to targeted therapies in patients with HER2-altered cancers, confirming previous findings of the predictive value of ctDNA in other cancer types.
“Such monitoring may be useful in clinical trials and eventually in practice,” she added.
Need for new methods to predict early tumor response
Limitations of standard radiographic tumor assessments present challenges in determining clinical response, particularly for patients receiving targeted therapies.
During her talk, Dr. Kurzrock explained that although targeted therapies are effective for patients with specific molecular alterations, standard imaging assessments fail to uncover molecular-level changes within tumors, limiting the ability of clinicians to accurately assess a patient’s response to targeted therapies.
“In addition to limitations with imaging, patients and physicians want to know as soon as possible whether or not the agents are effective, especially if there are side effects,” Dr. Kurzrock during an interview. She added that monitoring early response may be especially important across tumor types, as HER2 therapies are increasingly being considered in the pan-cancer setting.
Commenting on the potential use of this method in other cancer types with HER2 alterations, Pashtoon Murtaza Kasi, MD, MS, noted that since the study relied on a tumor-informed assay, it would be applicable across diverse tumor types.
“It is less about tissue type but more about that particular patient’s tumor at that instant in time for which a unique barcode is created,” said Dr. Kasi, a medical oncologist at Weill Cornell Medicine, New York, who was not involved in the study.
In an interview, he added that the shedding and biology would affect the assay’s performance for some tissue types.
Design of patient-specific ctDNA assays
In this retrospective study, the researchers examined ctDNA dynamics in 58 patients with various HER2-positive tumor types, including breast, colorectal, and other solid malignancies harboring HER2 alterations. All the patients received combination HER2-targeted therapy with trastuzumab and pertuzumab in the phase 2 basket trial My Pathway (NCT02091141).
By leveraging comprehensive genomic profiling of each patient’s tumor, the researchers designed personalized ctDNA assays, tracking 2-16 tumor-specific genetic variants in the patients’ blood samples. FoundationOne Tracker was used to detect and quantify ctDNA at baseline and the third cycle of therapy (cycle 3 day 1, or C3D1).
During an interview, Dr. Kurzrock explained that FoundationOne Tracker is a personalized ctDNA monitoring assay that allows for the detection of ctDNA in plasma, enabling ongoing liquid-based monitoring and highly sensitive quantification of ctDNA levels as mean tumor molecules per milliliter of plasma.
Among the 52 patients for whom personalized ctDNA assays were successfully designed, 48 (92.3%) had ctDNA data available at baseline, with a median of 100.7 tumor molecules per milliliter of plasma. Most patients (89.6%) were deemed ctDNA-positive, with a median of 119.5 tumor molecules per milliliter of plasma.
Changes in ctDNA levels predict patient survival
The researchers found that patients who experienced a greater than 90% decline in ctDNA levels by the third treatment cycle had significantly longer overall survival (OS) than those with less than 90% ctDNA decline or any increase. According to data presented by Dr. Kurzrock, the median OS was not reached in the group with greater than 90% decline in on-treatment ctDNA levels, versus 9.4 months in the group with less than 90% decline or ctDNA increase (P = .007). These findings held true when the analysis was limited to the 14 patients with colorectal cancer, in which median OS was not reached in the group with greater than 90% decline in on-treatment ctDNA levels, versus 10.2 months in the group with less than 90% decline or ctDNA increase (P = 0.04).
Notably, the prognostic significance of ctDNA changes remained even among patients exhibiting radiographic stable disease, underscoring the limitations of relying solely on anatomic tumor measurements and highlighting the potential for ctDNA monitoring to complement standard clinical assessments. In the subset of patients with radiographic stable disease, those with a greater than 90% ctDNA decline had significantly longer OS than those with less ctDNA reduction (not reached versus 9.4 months; P = .01).
“When used as a complement to imaging, tissue-informed ctDNA monitoring with FoundationOne Tracker can provide more accuracy than imaging alone,” Dr. Kurzrock noted in an interview.
Dr. Kasi echoed Dr. Kurzrock’s enthusiasm regarding the clinical usefulness of these findings, saying, “Not only can you see very early on in whom the ctDNA is going down and clearing, but you can also tell apart within the group who has ‘stable disease’ as to who is deriving more benefit.”
The researchers also observed that increases in on-treatment ctDNA levels often preceded radiographic evidence of disease progression by a median of 1.3 months. These findings highlight the potential for ctDNA monitoring to complement standard clinical assessments, allowing us to detect treatment response and disease progression earlier than what is possible with imaging alone, Dr. Kurzrock explained during her talk. “This early warning signal could allow clinicians to intervene and modify treatment strategies before overt clinical deterioration,” she said.
In an interview, Dr. Kasi highlighted that this high sensitivity and specificity and the short half-life of the tumor-informed ctDNA assay make this liquid biopsy of great clinical value. “The short half-life of a few hours means that if you do an intervention to treat cancer with HER2-directed therapy, you can very quickly assess response to therapy way earlier than traditional radiographic methods.”
Dr. Kasi cautioned, however, that this assay would not capture whether new mutations or HER2 loss occurred at the time of resistance. “A repeat tissue biopsy or a next-generation sequencing-based plasma-only assay would be required for that,” he said.
Implementation of ctDNA monitoring in clinical trials
Dr. Kurzrock acknowledged that further research is needed to validate these results in larger, prospective cohorts before FoundationOne Tracker is adopted in the clinic. She noted, however, that this retrospective analysis, along with results from previous studies, provides a rationale for the use of ctDNA monitoring in clinical trials.
“In some centers like ours, ctDNA monitoring is already part of our standard of care since not only does it help from a physician standpoint to have a more accurate and early assessment of response, but patients also appreciate the information gained from ctDNA dynamics,” Dr. Kasi said in an interview. He explained that when radiographic findings are equivocal, ctDNA monitoring is an additional tool in their toolbox to help guide care.
He noted, however, that the cost is a challenge for implementing ctDNA monitoring as a complementary tool for real-time treatment response monitoring. “For serial monitoring, helping to reduce costs would be important in the long run,” he said in an interview. He added that obtaining sufficient tissue for testing using a tumor-informed assay can present a logistical challenge, at least for the first test. “You need sufficient tissue to make the barcode that you then follow along,” he explained.
“Developing guidelines through systematic studies about testing cadence would also be important. This would help establish whether ctDNA monitoring is helpful,” Dr. Kasi said in an interview. He explained that in some situations, biological variables affect the shedding and detection of ctDNA beyond the assay — in those cases, ctDNA monitoring may not be helpful. “Like any test, it is not meant for every patient or clinical question,” Dr. Kasi concluded.
Dr. Kurzrock and Dr. Kasi reported no relationships with entities whose primary business is producing, marketing, selling, reselling, or distributing healthcare products used by or on patients.
This was the main finding of new data presented by study author Razelle Kurzrock, MD, at the American Association for Cancer Research annual meeting.
“We found that on-treatment ctDNA can detect progression before standard-of-care response assessments. These data suggest that monitoring ctDNA can provide clinicians with important prognostic information that may guide treatment decisions,” Dr. Kurzrock, professor at the Medical College of Wisconsin, Milwaukee, said during her presentation.
Commenting on the clinical implications of these findings during an interview, she said the results suggest that ctDNA dynamics provide an early window into predicting response to targeted therapies in patients with HER2-altered cancers, confirming previous findings of the predictive value of ctDNA in other cancer types.
“Such monitoring may be useful in clinical trials and eventually in practice,” she added.
Need for new methods to predict early tumor response
Limitations of standard radiographic tumor assessments present challenges in determining clinical response, particularly for patients receiving targeted therapies.
During her talk, Dr. Kurzrock explained that although targeted therapies are effective for patients with specific molecular alterations, standard imaging assessments fail to uncover molecular-level changes within tumors, limiting the ability of clinicians to accurately assess a patient’s response to targeted therapies.
“In addition to limitations with imaging, patients and physicians want to know as soon as possible whether or not the agents are effective, especially if there are side effects,” Dr. Kurzrock during an interview. She added that monitoring early response may be especially important across tumor types, as HER2 therapies are increasingly being considered in the pan-cancer setting.
Commenting on the potential use of this method in other cancer types with HER2 alterations, Pashtoon Murtaza Kasi, MD, MS, noted that since the study relied on a tumor-informed assay, it would be applicable across diverse tumor types.
“It is less about tissue type but more about that particular patient’s tumor at that instant in time for which a unique barcode is created,” said Dr. Kasi, a medical oncologist at Weill Cornell Medicine, New York, who was not involved in the study.
In an interview, he added that the shedding and biology would affect the assay’s performance for some tissue types.
Design of patient-specific ctDNA assays
In this retrospective study, the researchers examined ctDNA dynamics in 58 patients with various HER2-positive tumor types, including breast, colorectal, and other solid malignancies harboring HER2 alterations. All the patients received combination HER2-targeted therapy with trastuzumab and pertuzumab in the phase 2 basket trial My Pathway (NCT02091141).
By leveraging comprehensive genomic profiling of each patient’s tumor, the researchers designed personalized ctDNA assays, tracking 2-16 tumor-specific genetic variants in the patients’ blood samples. FoundationOne Tracker was used to detect and quantify ctDNA at baseline and the third cycle of therapy (cycle 3 day 1, or C3D1).
During an interview, Dr. Kurzrock explained that FoundationOne Tracker is a personalized ctDNA monitoring assay that allows for the detection of ctDNA in plasma, enabling ongoing liquid-based monitoring and highly sensitive quantification of ctDNA levels as mean tumor molecules per milliliter of plasma.
Among the 52 patients for whom personalized ctDNA assays were successfully designed, 48 (92.3%) had ctDNA data available at baseline, with a median of 100.7 tumor molecules per milliliter of plasma. Most patients (89.6%) were deemed ctDNA-positive, with a median of 119.5 tumor molecules per milliliter of plasma.
Changes in ctDNA levels predict patient survival
The researchers found that patients who experienced a greater than 90% decline in ctDNA levels by the third treatment cycle had significantly longer overall survival (OS) than those with less than 90% ctDNA decline or any increase. According to data presented by Dr. Kurzrock, the median OS was not reached in the group with greater than 90% decline in on-treatment ctDNA levels, versus 9.4 months in the group with less than 90% decline or ctDNA increase (P = .007). These findings held true when the analysis was limited to the 14 patients with colorectal cancer, in which median OS was not reached in the group with greater than 90% decline in on-treatment ctDNA levels, versus 10.2 months in the group with less than 90% decline or ctDNA increase (P = 0.04).
Notably, the prognostic significance of ctDNA changes remained even among patients exhibiting radiographic stable disease, underscoring the limitations of relying solely on anatomic tumor measurements and highlighting the potential for ctDNA monitoring to complement standard clinical assessments. In the subset of patients with radiographic stable disease, those with a greater than 90% ctDNA decline had significantly longer OS than those with less ctDNA reduction (not reached versus 9.4 months; P = .01).
“When used as a complement to imaging, tissue-informed ctDNA monitoring with FoundationOne Tracker can provide more accuracy than imaging alone,” Dr. Kurzrock noted in an interview.
Dr. Kasi echoed Dr. Kurzrock’s enthusiasm regarding the clinical usefulness of these findings, saying, “Not only can you see very early on in whom the ctDNA is going down and clearing, but you can also tell apart within the group who has ‘stable disease’ as to who is deriving more benefit.”
The researchers also observed that increases in on-treatment ctDNA levels often preceded radiographic evidence of disease progression by a median of 1.3 months. These findings highlight the potential for ctDNA monitoring to complement standard clinical assessments, allowing us to detect treatment response and disease progression earlier than what is possible with imaging alone, Dr. Kurzrock explained during her talk. “This early warning signal could allow clinicians to intervene and modify treatment strategies before overt clinical deterioration,” she said.
In an interview, Dr. Kasi highlighted that this high sensitivity and specificity and the short half-life of the tumor-informed ctDNA assay make this liquid biopsy of great clinical value. “The short half-life of a few hours means that if you do an intervention to treat cancer with HER2-directed therapy, you can very quickly assess response to therapy way earlier than traditional radiographic methods.”
Dr. Kasi cautioned, however, that this assay would not capture whether new mutations or HER2 loss occurred at the time of resistance. “A repeat tissue biopsy or a next-generation sequencing-based plasma-only assay would be required for that,” he said.
Implementation of ctDNA monitoring in clinical trials
Dr. Kurzrock acknowledged that further research is needed to validate these results in larger, prospective cohorts before FoundationOne Tracker is adopted in the clinic. She noted, however, that this retrospective analysis, along with results from previous studies, provides a rationale for the use of ctDNA monitoring in clinical trials.
“In some centers like ours, ctDNA monitoring is already part of our standard of care since not only does it help from a physician standpoint to have a more accurate and early assessment of response, but patients also appreciate the information gained from ctDNA dynamics,” Dr. Kasi said in an interview. He explained that when radiographic findings are equivocal, ctDNA monitoring is an additional tool in their toolbox to help guide care.
He noted, however, that the cost is a challenge for implementing ctDNA monitoring as a complementary tool for real-time treatment response monitoring. “For serial monitoring, helping to reduce costs would be important in the long run,” he said in an interview. He added that obtaining sufficient tissue for testing using a tumor-informed assay can present a logistical challenge, at least for the first test. “You need sufficient tissue to make the barcode that you then follow along,” he explained.
“Developing guidelines through systematic studies about testing cadence would also be important. This would help establish whether ctDNA monitoring is helpful,” Dr. Kasi said in an interview. He explained that in some situations, biological variables affect the shedding and detection of ctDNA beyond the assay — in those cases, ctDNA monitoring may not be helpful. “Like any test, it is not meant for every patient or clinical question,” Dr. Kasi concluded.
Dr. Kurzrock and Dr. Kasi reported no relationships with entities whose primary business is producing, marketing, selling, reselling, or distributing healthcare products used by or on patients.
This was the main finding of new data presented by study author Razelle Kurzrock, MD, at the American Association for Cancer Research annual meeting.
“We found that on-treatment ctDNA can detect progression before standard-of-care response assessments. These data suggest that monitoring ctDNA can provide clinicians with important prognostic information that may guide treatment decisions,” Dr. Kurzrock, professor at the Medical College of Wisconsin, Milwaukee, said during her presentation.
Commenting on the clinical implications of these findings during an interview, she said the results suggest that ctDNA dynamics provide an early window into predicting response to targeted therapies in patients with HER2-altered cancers, confirming previous findings of the predictive value of ctDNA in other cancer types.
“Such monitoring may be useful in clinical trials and eventually in practice,” she added.
Need for new methods to predict early tumor response
Limitations of standard radiographic tumor assessments present challenges in determining clinical response, particularly for patients receiving targeted therapies.
During her talk, Dr. Kurzrock explained that although targeted therapies are effective for patients with specific molecular alterations, standard imaging assessments fail to uncover molecular-level changes within tumors, limiting the ability of clinicians to accurately assess a patient’s response to targeted therapies.
“In addition to limitations with imaging, patients and physicians want to know as soon as possible whether or not the agents are effective, especially if there are side effects,” Dr. Kurzrock during an interview. She added that monitoring early response may be especially important across tumor types, as HER2 therapies are increasingly being considered in the pan-cancer setting.
Commenting on the potential use of this method in other cancer types with HER2 alterations, Pashtoon Murtaza Kasi, MD, MS, noted that since the study relied on a tumor-informed assay, it would be applicable across diverse tumor types.
“It is less about tissue type but more about that particular patient’s tumor at that instant in time for which a unique barcode is created,” said Dr. Kasi, a medical oncologist at Weill Cornell Medicine, New York, who was not involved in the study.
In an interview, he added that the shedding and biology would affect the assay’s performance for some tissue types.
Design of patient-specific ctDNA assays
In this retrospective study, the researchers examined ctDNA dynamics in 58 patients with various HER2-positive tumor types, including breast, colorectal, and other solid malignancies harboring HER2 alterations. All the patients received combination HER2-targeted therapy with trastuzumab and pertuzumab in the phase 2 basket trial My Pathway (NCT02091141).
By leveraging comprehensive genomic profiling of each patient’s tumor, the researchers designed personalized ctDNA assays, tracking 2-16 tumor-specific genetic variants in the patients’ blood samples. FoundationOne Tracker was used to detect and quantify ctDNA at baseline and the third cycle of therapy (cycle 3 day 1, or C3D1).
During an interview, Dr. Kurzrock explained that FoundationOne Tracker is a personalized ctDNA monitoring assay that allows for the detection of ctDNA in plasma, enabling ongoing liquid-based monitoring and highly sensitive quantification of ctDNA levels as mean tumor molecules per milliliter of plasma.
Among the 52 patients for whom personalized ctDNA assays were successfully designed, 48 (92.3%) had ctDNA data available at baseline, with a median of 100.7 tumor molecules per milliliter of plasma. Most patients (89.6%) were deemed ctDNA-positive, with a median of 119.5 tumor molecules per milliliter of plasma.
Changes in ctDNA levels predict patient survival
The researchers found that patients who experienced a greater than 90% decline in ctDNA levels by the third treatment cycle had significantly longer overall survival (OS) than those with less than 90% ctDNA decline or any increase. According to data presented by Dr. Kurzrock, the median OS was not reached in the group with greater than 90% decline in on-treatment ctDNA levels, versus 9.4 months in the group with less than 90% decline or ctDNA increase (P = .007). These findings held true when the analysis was limited to the 14 patients with colorectal cancer, in which median OS was not reached in the group with greater than 90% decline in on-treatment ctDNA levels, versus 10.2 months in the group with less than 90% decline or ctDNA increase (P = 0.04).
Notably, the prognostic significance of ctDNA changes remained even among patients exhibiting radiographic stable disease, underscoring the limitations of relying solely on anatomic tumor measurements and highlighting the potential for ctDNA monitoring to complement standard clinical assessments. In the subset of patients with radiographic stable disease, those with a greater than 90% ctDNA decline had significantly longer OS than those with less ctDNA reduction (not reached versus 9.4 months; P = .01).
“When used as a complement to imaging, tissue-informed ctDNA monitoring with FoundationOne Tracker can provide more accuracy than imaging alone,” Dr. Kurzrock noted in an interview.
Dr. Kasi echoed Dr. Kurzrock’s enthusiasm regarding the clinical usefulness of these findings, saying, “Not only can you see very early on in whom the ctDNA is going down and clearing, but you can also tell apart within the group who has ‘stable disease’ as to who is deriving more benefit.”
The researchers also observed that increases in on-treatment ctDNA levels often preceded radiographic evidence of disease progression by a median of 1.3 months. These findings highlight the potential for ctDNA monitoring to complement standard clinical assessments, allowing us to detect treatment response and disease progression earlier than what is possible with imaging alone, Dr. Kurzrock explained during her talk. “This early warning signal could allow clinicians to intervene and modify treatment strategies before overt clinical deterioration,” she said.
In an interview, Dr. Kasi highlighted that this high sensitivity and specificity and the short half-life of the tumor-informed ctDNA assay make this liquid biopsy of great clinical value. “The short half-life of a few hours means that if you do an intervention to treat cancer with HER2-directed therapy, you can very quickly assess response to therapy way earlier than traditional radiographic methods.”
Dr. Kasi cautioned, however, that this assay would not capture whether new mutations or HER2 loss occurred at the time of resistance. “A repeat tissue biopsy or a next-generation sequencing-based plasma-only assay would be required for that,” he said.
Implementation of ctDNA monitoring in clinical trials
Dr. Kurzrock acknowledged that further research is needed to validate these results in larger, prospective cohorts before FoundationOne Tracker is adopted in the clinic. She noted, however, that this retrospective analysis, along with results from previous studies, provides a rationale for the use of ctDNA monitoring in clinical trials.
“In some centers like ours, ctDNA monitoring is already part of our standard of care since not only does it help from a physician standpoint to have a more accurate and early assessment of response, but patients also appreciate the information gained from ctDNA dynamics,” Dr. Kasi said in an interview. He explained that when radiographic findings are equivocal, ctDNA monitoring is an additional tool in their toolbox to help guide care.
He noted, however, that the cost is a challenge for implementing ctDNA monitoring as a complementary tool for real-time treatment response monitoring. “For serial monitoring, helping to reduce costs would be important in the long run,” he said in an interview. He added that obtaining sufficient tissue for testing using a tumor-informed assay can present a logistical challenge, at least for the first test. “You need sufficient tissue to make the barcode that you then follow along,” he explained.
“Developing guidelines through systematic studies about testing cadence would also be important. This would help establish whether ctDNA monitoring is helpful,” Dr. Kasi said in an interview. He explained that in some situations, biological variables affect the shedding and detection of ctDNA beyond the assay — in those cases, ctDNA monitoring may not be helpful. “Like any test, it is not meant for every patient or clinical question,” Dr. Kasi concluded.
Dr. Kurzrock and Dr. Kasi reported no relationships with entities whose primary business is producing, marketing, selling, reselling, or distributing healthcare products used by or on patients.
FROM AACR 2024

