User login
Hand, foot, and mouth disease: Identifying and managing an acute viral syndrome
Hand, foot, and mouth disease (HFMD) is typically a benign childhood infection—except when it isn’t so benign or when it occurs in adults.
The usual presentation is in a child with fever, oral ulcerations, and papules on the palms of the hands and the soles of the feet.1 However, severe complications can occur, including central nervous system involvement and cardiopulmonary failure, and can lead to significant morbidity and even death.2 Fortunately, these complications are rare.
Less common in North America than in other regions, HFMD has recurrently broken out in many areas of Southern Asia and the surrounding Pacific region. However, several North American outbreaks have been documented in recent years and have affected unexpected numbers of immunocompetent adults, demonstrating that this disease is of worldwide importance in adults as well as children.3
Because HFMD has the potential to reach epidemic levels in the United States, early recognition is paramount, and primary care physicians need to be familiar with its common signs and symptoms.
USUALLY A SUMMER DISEASE
HFMD occurs all around the world, exhibiting seasonal variation in temperate climates. In these locations, individual cases and regional outbreaks usually occur in the spring, summer, and fall. No sexual predisposition has been documented. Most symptomatic cases are in children under the age of 10.
OUTBREAKS AROUND THE WORLD
The disease was first described more than 40 years ago, with several large outbreaks in the last 16 years.
1998—An outbreak in Taiwan affected more than 1.5 million people, mostly children. Severe cases numbered just over 400, and 78 children died.4
2008—China,5 Singapore,6 Vietnam,7 Mongolia,8 and Brunei9 were stricken with an outbreak that affected 30,000 people and led to more than 50 deaths.
2009—An outbreak in the Henan and Shandong provinces of eastern China killed 35 people.10
2010—In several southern Chinese regions, more than 70,000 people were infected, with almost 600 deaths.11
2011 to the present. The United States has had several outbreaks in the last 3 years. Although HFMD is not one of the diseases that must be reported to public health authorities in the United States, from November 2011 to February 2012 the US Centers for Disease Control and Prevention (CDC) received reports of 63 possible cases: 38 in Alabama, 17 in Nevada, 7 in California, and 1 in Connecticut.1 Fifteen of the patients were adults, and more than half had contacts who were sick.
The most recent US outbreak, in Alabama,12 was atypical because it occurred in the winter.
CAUSED BY ENTEROVIRUSES
HFMD is caused by infection with a variety of viruses in the genus Enterovirus, a large group that in turn is part of the larger Picornaviridae family.13 The taxonomy of this genus is complicated and subject to revision; species include coxsackieviruses, polioviruses, enteroviruses, and echoviruses. They are all small, nonenveloped, single-stranded RNA viruses.
The most common strains that cause HFMD are coxsackievirus A16 and enterovirus 71. In addition, coxsackievirus A6 may be emerging, and many other coxsackievirus strains have been directly implicated, including A5, A7, A9, A10, B2, and B5.
Coxsackievirus A16 is the leading cause of HFMD.
Enterovirus 71 is the second most common cause of HFMD and has also caused outbreaks. It usually results in benign disease. However, among the causes of HFMD, it is associated with more prominent central nervous system involvement14 and is the most common cause of viral meningoencephalitis in children.
Coxsackievirus A6. In December 2011, the California Department of Public Health isolated a strain of coxsackievirus A6 that caused extensive rash and nail shedding.15 Among the 63 possible cases of HFMD reported to the CDC from November 2011 to February 2012, specimens for clinical testing were obtained in 34, and 25 of those demonstrated coxsackievirus A6 infection.3
FEVER, ORAL ULCERS, RASH ON HANDS AND FEET
The typical clinical manifestations of HFMD are fever, stomatitis with oral ulcers, and an exanthem affecting the palms, soles, and other parts of the body. These last less than 7 to 10 days, usually occur during the spring to fall months, and have a benign course.
The incubation period is 3 to 5 days, with a prodrome that may include fever, malaise, abdominal pain, and myalgia before the onset of oral and cutaneous findings. Painful oral ulcers may precede the exanthem and can result in dehydration.16
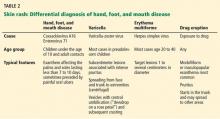
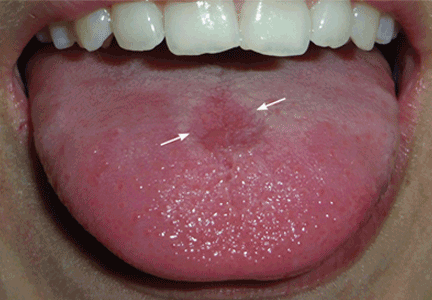
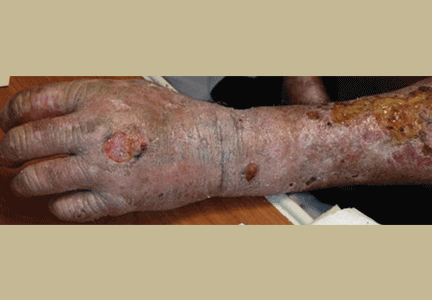
The cutaneous manifestation of HFMD is typically a papulovesicular rash affecting the palms, soles, and buttocks (Figure 1). Other sites may include the knees, elbows, and the dorsal surfaces of the hands and feet. The lesions may be maculopapular and can be either asymptomatic or tender and painful. Desquamation can follow the exanthem, and lesions usually resolve without scarring or secondary infection.16,17
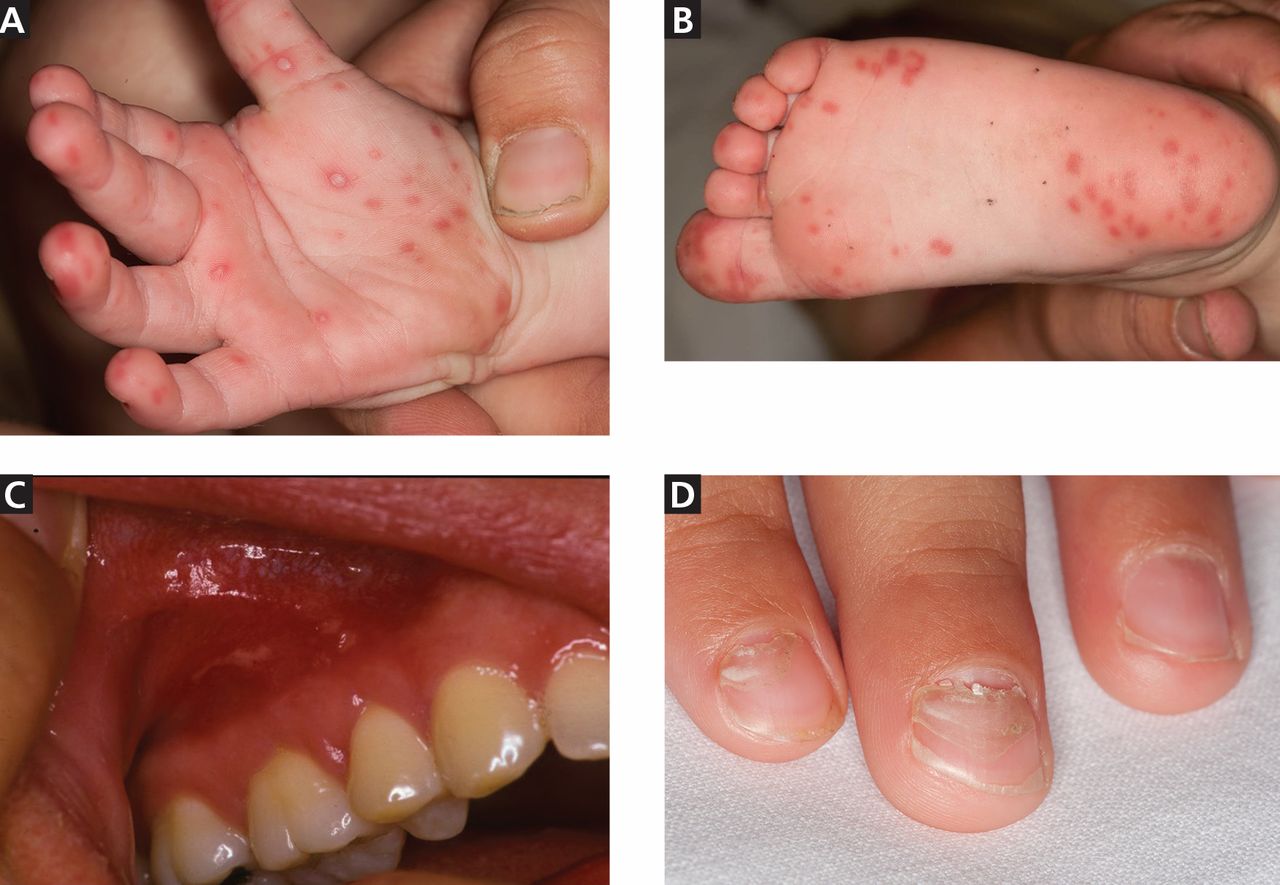
Table 1 and Table 2 compare HFMD with other common illnesses that can cause similar skin and mucosal findings. In particular, herpangina has the identical clinical presentation as HFMD except that it does not cause skin lesions. It is caused by many of the same enteroviruses linked to HFMD.
Different viruses, different signs?
The numerous viruses that cause HFMD usually cause similar signs and symptoms during bouts of typical, self-limited disease. However, neurologic and cardiopulmonary involvement, which are fortunately rare, are more often associated with enterovirus 71 infection.
Nail manifestations are common in HFMD. Nail separation from the nail matrix (onychomadesis) was associated with coxsackievirus A6 infection during a 2010 outbreak of HFMD in Taiwan and in a 2009 outbreak in Finland.18 Moreover, this virus was cultured from a nail specimen in one patient, suggesting viral infiltration as the cause of nail-matrix arrest.19
Perioral skin eruptions, desquamation, and Beau lines have also been associated with coxsackievirus A6.18 Beau lines are transverse depressions of the nail, most evident in the central nail plate; when seen on multiple nails, they imply a systemic illness causing disruption of nail matrix growth.20
Atypical HFMD and coxsackievirus A6
Atypical HFMD has recently been described in connection with coxsackievirus A6. Lott et al21 reported five cases of coxsackievirus A6-associated HFMD in 2013. Atypically, three of the affected patients presented in winter months, two were adults, and two had widespread skin involvement.21
Mathes et al22 reported a series of 80 cases of enteroviral infections in which the lesions had a predilection for the antecubital and popliteal fossae and were similar in severity and distribution to those seen in eczema herpeticum or Kaposi varicelliform eruption in patients with and patients without a history of atopic dermatitis. They named this find-clinical finding of pronounced coxsackievirus-associated skin disease at sites previously affected by atopic dermatitis.
Additional cutaneous findings of coxsackievirus A6 infection may include onychomadesis, Beau lines, and vesiculobullous lesions. Patients with atypical, coxsackievirus A6-associated HFMD may not have oral lesions.23
In the five cases reported by Lott et al,21 significant systemic symptoms (fever, chills, diarrhea, and myalgias) led all but one of the patients to seek care in an emergency department. However, atypical HFMD has not been associated with life-threatening illness.
Atypical HFMD associated with coxsackievirus A6 is an emerging entity in the United States, and the acuity of both cutaneous and systemic symptoms poses a diagnostic dilemma. Furthermore, infection has been documented in immunocompetent adults.23 Familiarity with the clinical findings may expedite appropriate care, prevent spread to contacts, and avoid unnecessary testing.
Neurologic and cardiopulmonary manifestations
Enteroviruses are the most common causes of viral meningoencephalitis in the United States. They mainly affect children and cause serious and potentially chronic disease in those with humoral immunodeficiencies.24 Neurologic and cardiopulmonary manifestations of HFMD are varied and extremely rare in the United States but should always be viewed clinically as signs of concern and severe disease.
Signs of potentially fatal disease that have been observed in young children include tachycardia, tachypnea, hypotension, hypertension, gastrointestinal bleeding, neurologic symptoms, leukocytosis, absence of oral lesions, and vomiting.2 Signs of dysautonomia, myoclonus, ataxia, and brainstem involvement may portend fatal disease in which rapid decompensation is the result of cardiogenic shock due to loss of ventricular contractility, causing pulmonary edema and end-organ dysfunction.16
Neurologic manifestations associated with enterovirus 71 infection include aseptic meningitis, a poliomyelitis-like syndrome, brainstem encephalitis, neurogenic pulmonary edema, opsoclonus-myoclonus syndrome, cerebellar ataxia, Guillain-Barré syndrome, and transverse myelitis.
Because some patients who have neurologic disease respond to treatment with high-dose methylprednisolone and intravenous immune globulin, there is reason to suspect that an autoimmune phenomenon triggered by the culprit enterovirus may be the cause of many of the neurologic symptoms.25
A 2012 meta-analysis26 found that an elevated white blood cell count and hyperglycemia could be clinically useful in distinguishing benign from severe HFMD. In patients with benign HFMD, white blood cell counts and blood glucose values were no different from those in healthy controls.26
DIAGNOSIS IS USUALLY CLINICAL
Most enteroviral infections are asymptomatic, but HFMD is a possibility if a patient has mild illness, fever, and a maculopapular or vesicular rash on the palms of the hands and soles of the feet, sometimes associated with oral ulcers (herpangina). Skin lesions can also be found on the legs, face, buttocks, and trunk.
In the United States, HFMD most commonly occurs in children under age 4 and is usually caused by coxsackievirus A16. Adults can also be affected, especially if they were in contact with children in child care, which was the case in approximately half of nonpediatric patients who tested positive for HFMD during an outbreak in several states between November 2011 and February 2012.3
The clinical characteristics of HFMD caused by enterovirus 71 may be somewhat different, with smaller vesicles, diffuse erythema of the trunk and limbs, and higher fever (temperature ≥ 39°C [102.2°F] for more than 3 days).27 However, the rash of coxsackievirus A16 HFMD may be more extensive and severe.
Other clinical manifestations of HFMD include nail dystrophies such as Beau lines and nail shedding, hyperglycemia, dehydration, and more serious and potentially life-threatening complications such as pulmonary edema28 and viral meningoencephalitis.29
Laboratory testing
In mild cases of HFMD, particularly in patients with a high probability of having the disease based on their clinical characteristics and sick contacts, laboratory testing is not necessary. Testing is usually reserved for severe cases and public health investigation of outbreaks.
Viral culture is the gold standard for diagnosing HFMD, but the final results can take nearly a week.
Polymerase chain reaction testing is faster, with a turnaround time of less than 1 day. It identifies viral RNA and is highly sensitive for detecting central nervous system infection.30
Where should samples be collected? Serum viremia precedes invasion of the skin and mucous membranes, so plasma can be tested. Inside the body, enteroviruses initially replicate in the gastrointestinal tract, although collecting a rectal swab or a stool sample is somewhat invasive. Further, in an enterovirus 71 epidemic in Taiwan, 93% of the patients had positive throat swabs, but only 30% tested positive by rectal swabs or analysis of the feces.27 At present, throat and vesicle specimens are considered to be the most useful sources for diagnostic purposes.16
ELISAs. Newly developed IgM-capture enzyme-linked immunosorbent assays (ELISAs) for coxsackievirus A16 and enterovirus 71 appear quite promising for diagnosing HFMD. These tests are inexpensive and detect IgM antibodies early and in a high percentage of patients. In the first week of the disease, the IgM detection rate was found to be 90.2% for enterovirus 71 and 68% for coxsackievirus A16.31
Cross-reactivity between these two viruses was a problem with ELISA testing in the past, causing false-positive results for enterovirus 71 in patients who in fact had coxsackievirus A16. The problem appears to be resolved in new versions that use specific enterovirus 71 proteins, eg, VP1.32
RECOGNITION AND PREVENTION ARE THE BEST MEDICINE
Recognizing HFMD early is crucial, because making the clinical diagnosis can identify patients who have signs of severe disease and can help protect future contacts and decrease the risk of an epidemic.
Infected patients continue to shed the virus for a long time, making hand hygiene and environmental control measures in health care settings and daycare centers of vital importance, to prevent spread of the infection.
Enteroviruses are stable in the environment and therefore capable of fecal-oral and oral-oral transmission. Humans are the only known natural hosts. No chemoprophylaxis or vaccination has been established to prevent HFMD. The recurrence of large-scale epidemics in the developing world is perhaps explained by ineffective sewage treatment and limited access to clean drinking water, especially in light of the fecal-oral spread of the virus. Intrafamilial spread of HFMD has been shown to be an important means of disease transmission, and asymptomatic adult carriers of these viruses may spread it to young children.33
The different viruses that cause HFMD result in a similar clinical presentation in most patients. Therefore, identifying HFMD caused by enterovirus 71, which carries a risk of severe and even fatal disease in young children vs a virus such as coxsackievirus A16, can be very difficult in practice without virologic testing. Thus, when diagnosed with HFMD, patients should be counseled to control all variables that could lead to further spread of the disease.
An analysis of epidemics in Asia suggested that public health awareness may have averted deaths in successive epidemics, highlighting the need to identify HFMD epidemics in communities and to educate patients and families about measures to prevent further spread of the virus in addition to standard supportive care.34
The CDC recommends35:
- Frequent hand-washing after toileting and changing diapers
- Disinfecting frequently used surfaces and objects, including toys
- Avoiding close contact with infected individuals and sharing of personal items such as utensils and cups.
These measures should be recommended to all affected patients.35
NO PROVEN ANTIVIRAL TREATMENT
No proven antiviral treatment exists for HFMD. Thus, the goals of treatment are typically supportive, as for any self-limited viral syndrome.16
Does acyclovir help? Shelley et al36 treated 13 patients (12 children and 1 adult) with acyclovir within 1 to 2 days of the onset of the HFMD rash and reported that it was beneficial, with significant relief of fever and skin lesions within 24 hours of starting therapy. These anecdotal results have not been replicated, and acyclovir is not an established treatment for HFMD.
If acyclovir does help, how does it work? Acyclovir, like other common antiviral medications, inactivates thymidine kinase, an enzyme produced by herpesviruses but not by HFMD-causing viruses like coxsackievirus A16. Shelley et al proposed that acyclovir may enhance the antiviral effect of the patient’s own interferon.36
Intravenous immunoglobulin has been used in severe cases during outbreaks in Asia, with retrospective data showing a potential ability to halt disease progression if used before the development of cardiopulmonary failure. However, this has not been studied prospectively and is not currently recommended.16
Acknowledgment: We would like to thank Dr. Salvador Alvarez of the Mayo Clinic Department of Infectious Disease and Dr. Donald Lookingbill of the Mayo Clinic Department of Dermatology for their collaboration.
- Centers for Disease Control and Prevention (CDC). Hand, Foot, and Mouth Disease (HFMD). www.cdc.gov/hand-foot-mouth/index.html. Accessed June 10, 2014.
- Chatproedprai S, Theanboonlers A, Korkong S, Thongmee C, Wananukul S, Poovorawan Y. Clinical and molecular characterization of hand-foot-and-mouth disease in Thailand, 2008–2009. Jpn J Infect Dis 2010; 63:229–233.
- Centers for Disease Control and Prevention (CDC). Notes from the field: severe hand, foot, and mouth disease associated with coxsackievirus A6—Alabama, Connecticut, California, and Nevada, November 2011–February 2012. MMWR Morb Mortal Wkly Rep 2012; 61:213–214.
- Ho M, Chen ER, Hsu KH, et al. An epidemic of enterovirus 71 infection in Taiwan. Taiwan Enterovirus Epidemic Working Group. N Engl J Med 1999; 341:929–935.
- BBC News. China virus toll continues rise. May 5, 2008. http://news.bbc.co.uk/2/hi/asia-pacific/7383796.stm. Accessed February 5, 2014.
- Suhaimi ND. HFMD: 1,000 cases a week in Singapore is unusual, says doc. Straits Times April 20, 2008.
- Viet Nam News: HFMD cases prompt tighter health screening at airport. May 15, 2008.
- UBPOST. EV-71 virus continues dramatic rise. May 22, 2008.
- Begawan BS. 1,053 HFMD cases recorded. Brunei Times. November 7, 2008.
- Chinaview. Hand-foot-mouth disease death toll rises to 17 in East China’s Shandong Province. April 9, 2009.
- Chinaview. China reports 537 deaths from hand-foot-mouth disease this year. June 24, 2010.
- Wolfson H. Outbreak of hand, foot and mouth disease severe in Alabama. Birmingham News February 13, 2012.
- Centers for Disease Control and Prevention (CDC). Non-Polio Enterovirus Infections. www.cdc.gov/non-polio-enterovirus/. Accessed June 10, 2014.
- Chan KP, Goh KT, Chong CY, Teo ES, Lau G, Ling AE. Epidemic hand, foot and mouth disease caused by human enterovirus 71, Singapore. Emerg Infect Dis 2003; 9:78–85.
- California Department of Public Health. Coxsackievirus A6 (CVA6). 2011. www.cdph.ca.gov/programs/cder/Pages/CVA6.aspx. Accessed June 10, 2014.
- World Health Organization: Western Pacific Region. A Guide to Clinical management and Public Health Response for Hand, Foot, and Mouth Disease (HFMD).
- Shin JU, Oh SH, Lee JH. A case of hand-foot-mouth disease in an immunocompetent adult. Ann Dermatol 2010; 22:216–218.
- Wei SH, Huang YP, Liu MC, et al. An outbreak of coxsackievirus A6 hand, foot, and mouth disease associated with onychomadesis in Taiwan, 2010. BMC Infect Dis 2011; 11:346.
- Osterback R, Vuorinen T, Linna M, Susi P, Hyypiä T, Waris M. Coxsackievirus A6 and hand, foot, and mouth disease, Finland. Emerg Infect Dis 2009; 15:1485–1488.
- Tosti A, Piraccini BM. Nail Disorders. In:Bolognia JL, Jorizzo JL, Schaffer JV, editors. Dermatology JV. 3rded. Elsevier Limited; 2012:1129–1144.
- Lott JP, Liu K, Landry ML, et al. Atypical hand-foot-and-mouth disease associated with coxsackievirus A6 infection. J Am Acad Dermatol 2013; 69:736–741.
- Mathes EF, Oza V, Frieden IJ, et al. ”Eczema coxsackium” and unusual cutaneous findings in an enterovirus outbreak. Pediatrics 2013; 132:e149–e157.
- Kaminska K, Martinetti G, Lucchini R, Kaya G, Mainetti C. Coxsackievirus A6 and hand, foot and mouth disease: three case reports of familial child-to-immunocompetent adult transmission and a literature review. Case Rep Dermatol 2013; 5:203–209.
- Romero JR. Diagnosis and management of enteroviral infections of the central nervous system. Curr Infect Dis Rep 2002; 4:309–316.
- Akiyama K, Imazeki R, Yoshii F, Koide T, Muto J. An adult case of hand, foot, and mouth disease caused by enterovirus 71 accompanied by opsoclonus myoclonica. Tokai J Exp Clin Med 2008; 33:143–145.
- Li Y, Zhu R, Qian Y, Deng J. The characteristics of blood glucose and WBC counts in peripheral blood of cases of hand foot and mouth disease in China: a systematic review. PLoS One 2012; 7:e29003.
- Chang LY, King CC, Hsu KH, et al. Risk factors of enterovirus 71 infection and associated hand, foot, and mouth disease/herpangina in children during an epidemic in Taiwan. Pediatrics 2002; 109:e88.
- Wang SM, Liu CC, Tseng HW, et al. Clinical spectrum of enterovirus 71 infection in children in southern Taiwan, with an emphasis on neurological complications. Clin Infect Dis 1999; 29:184–190.
- Chang LY, Lin TY, Hsu KH, et al. Clinical features and risk factors of pulmonary oedema after enterovirus-71-related hand, foot, and mouth disease. Lancet 1999; 354:1682–1686.
- Mayo Clinic Laboratories. Enterovirus, Molecular Detection, PCR, Plasma. www.mayomedicallaboratories.com/test-catalog/Clinical+and+Interpretive/89893. Accessed June 10, 2014.
- Yu N, Guo M, He SJ, et al. Evaluation of human enterovirus 71 and coxsackievirus A16 specific immunoglobulin M antibodies for diagnosis of hand-foot-and-mouth disease. Virol J 2012; 9:12.
- Wang C, You A, Tian X, et al. Analysis and solution of false-positives when testing CVA16 sera using an antibody assay against the EV71 virus. Virus Res 2013; 176:33–36.
- Liu MY, Liu W, Luo J, et al. Characterization of an outbreak of hand, foot, and mouth disease in Nanchang, China in 2010. PLoS One 2011; 6:e25287.
- Zhang J, Sun J, Chang Z, Zhang W, Wang Z, Feng Z. Characterization of hand, foot, and mouth disease in China between 2008 and 2009. Biomed Environ Sci 2011; 24:214–221.
- Centers for Disease Control and Prevention (CDC). Hand, Foot, and Mouth Disease: Prevention & Treatment. www.cdc.gov/hand-foot-mouth/about/prevention-treatment.html. Accessed June 10, 2014.
- Shelley WB, Hashim M, Shelley ED. Acyclovir in the treatment of hand-foot-and-mouth disease. Cutis 1996; 57:232–234.
Hand, foot, and mouth disease (HFMD) is typically a benign childhood infection—except when it isn’t so benign or when it occurs in adults.
The usual presentation is in a child with fever, oral ulcerations, and papules on the palms of the hands and the soles of the feet.1 However, severe complications can occur, including central nervous system involvement and cardiopulmonary failure, and can lead to significant morbidity and even death.2 Fortunately, these complications are rare.
Less common in North America than in other regions, HFMD has recurrently broken out in many areas of Southern Asia and the surrounding Pacific region. However, several North American outbreaks have been documented in recent years and have affected unexpected numbers of immunocompetent adults, demonstrating that this disease is of worldwide importance in adults as well as children.3
Because HFMD has the potential to reach epidemic levels in the United States, early recognition is paramount, and primary care physicians need to be familiar with its common signs and symptoms.
USUALLY A SUMMER DISEASE
HFMD occurs all around the world, exhibiting seasonal variation in temperate climates. In these locations, individual cases and regional outbreaks usually occur in the spring, summer, and fall. No sexual predisposition has been documented. Most symptomatic cases are in children under the age of 10.
OUTBREAKS AROUND THE WORLD
The disease was first described more than 40 years ago, with several large outbreaks in the last 16 years.
1998—An outbreak in Taiwan affected more than 1.5 million people, mostly children. Severe cases numbered just over 400, and 78 children died.4
2008—China,5 Singapore,6 Vietnam,7 Mongolia,8 and Brunei9 were stricken with an outbreak that affected 30,000 people and led to more than 50 deaths.
2009—An outbreak in the Henan and Shandong provinces of eastern China killed 35 people.10
2010—In several southern Chinese regions, more than 70,000 people were infected, with almost 600 deaths.11
2011 to the present. The United States has had several outbreaks in the last 3 years. Although HFMD is not one of the diseases that must be reported to public health authorities in the United States, from November 2011 to February 2012 the US Centers for Disease Control and Prevention (CDC) received reports of 63 possible cases: 38 in Alabama, 17 in Nevada, 7 in California, and 1 in Connecticut.1 Fifteen of the patients were adults, and more than half had contacts who were sick.
The most recent US outbreak, in Alabama,12 was atypical because it occurred in the winter.
CAUSED BY ENTEROVIRUSES
HFMD is caused by infection with a variety of viruses in the genus Enterovirus, a large group that in turn is part of the larger Picornaviridae family.13 The taxonomy of this genus is complicated and subject to revision; species include coxsackieviruses, polioviruses, enteroviruses, and echoviruses. They are all small, nonenveloped, single-stranded RNA viruses.
The most common strains that cause HFMD are coxsackievirus A16 and enterovirus 71. In addition, coxsackievirus A6 may be emerging, and many other coxsackievirus strains have been directly implicated, including A5, A7, A9, A10, B2, and B5.
Coxsackievirus A16 is the leading cause of HFMD.
Enterovirus 71 is the second most common cause of HFMD and has also caused outbreaks. It usually results in benign disease. However, among the causes of HFMD, it is associated with more prominent central nervous system involvement14 and is the most common cause of viral meningoencephalitis in children.
Coxsackievirus A6. In December 2011, the California Department of Public Health isolated a strain of coxsackievirus A6 that caused extensive rash and nail shedding.15 Among the 63 possible cases of HFMD reported to the CDC from November 2011 to February 2012, specimens for clinical testing were obtained in 34, and 25 of those demonstrated coxsackievirus A6 infection.3
FEVER, ORAL ULCERS, RASH ON HANDS AND FEET
The typical clinical manifestations of HFMD are fever, stomatitis with oral ulcers, and an exanthem affecting the palms, soles, and other parts of the body. These last less than 7 to 10 days, usually occur during the spring to fall months, and have a benign course.
The incubation period is 3 to 5 days, with a prodrome that may include fever, malaise, abdominal pain, and myalgia before the onset of oral and cutaneous findings. Painful oral ulcers may precede the exanthem and can result in dehydration.16
The cutaneous manifestation of HFMD is typically a papulovesicular rash affecting the palms, soles, and buttocks (Figure 1). Other sites may include the knees, elbows, and the dorsal surfaces of the hands and feet. The lesions may be maculopapular and can be either asymptomatic or tender and painful. Desquamation can follow the exanthem, and lesions usually resolve without scarring or secondary infection.16,17

Table 1 and Table 2 compare HFMD with other common illnesses that can cause similar skin and mucosal findings. In particular, herpangina has the identical clinical presentation as HFMD except that it does not cause skin lesions. It is caused by many of the same enteroviruses linked to HFMD.
Different viruses, different signs?
The numerous viruses that cause HFMD usually cause similar signs and symptoms during bouts of typical, self-limited disease. However, neurologic and cardiopulmonary involvement, which are fortunately rare, are more often associated with enterovirus 71 infection.
Nail manifestations are common in HFMD. Nail separation from the nail matrix (onychomadesis) was associated with coxsackievirus A6 infection during a 2010 outbreak of HFMD in Taiwan and in a 2009 outbreak in Finland.18 Moreover, this virus was cultured from a nail specimen in one patient, suggesting viral infiltration as the cause of nail-matrix arrest.19
Perioral skin eruptions, desquamation, and Beau lines have also been associated with coxsackievirus A6.18 Beau lines are transverse depressions of the nail, most evident in the central nail plate; when seen on multiple nails, they imply a systemic illness causing disruption of nail matrix growth.20
Atypical HFMD and coxsackievirus A6
Atypical HFMD has recently been described in connection with coxsackievirus A6. Lott et al21 reported five cases of coxsackievirus A6-associated HFMD in 2013. Atypically, three of the affected patients presented in winter months, two were adults, and two had widespread skin involvement.21
Mathes et al22 reported a series of 80 cases of enteroviral infections in which the lesions had a predilection for the antecubital and popliteal fossae and were similar in severity and distribution to those seen in eczema herpeticum or Kaposi varicelliform eruption in patients with and patients without a history of atopic dermatitis. They named this find-clinical finding of pronounced coxsackievirus-associated skin disease at sites previously affected by atopic dermatitis.
Additional cutaneous findings of coxsackievirus A6 infection may include onychomadesis, Beau lines, and vesiculobullous lesions. Patients with atypical, coxsackievirus A6-associated HFMD may not have oral lesions.23
In the five cases reported by Lott et al,21 significant systemic symptoms (fever, chills, diarrhea, and myalgias) led all but one of the patients to seek care in an emergency department. However, atypical HFMD has not been associated with life-threatening illness.
Atypical HFMD associated with coxsackievirus A6 is an emerging entity in the United States, and the acuity of both cutaneous and systemic symptoms poses a diagnostic dilemma. Furthermore, infection has been documented in immunocompetent adults.23 Familiarity with the clinical findings may expedite appropriate care, prevent spread to contacts, and avoid unnecessary testing.
Neurologic and cardiopulmonary manifestations
Enteroviruses are the most common causes of viral meningoencephalitis in the United States. They mainly affect children and cause serious and potentially chronic disease in those with humoral immunodeficiencies.24 Neurologic and cardiopulmonary manifestations of HFMD are varied and extremely rare in the United States but should always be viewed clinically as signs of concern and severe disease.
Signs of potentially fatal disease that have been observed in young children include tachycardia, tachypnea, hypotension, hypertension, gastrointestinal bleeding, neurologic symptoms, leukocytosis, absence of oral lesions, and vomiting.2 Signs of dysautonomia, myoclonus, ataxia, and brainstem involvement may portend fatal disease in which rapid decompensation is the result of cardiogenic shock due to loss of ventricular contractility, causing pulmonary edema and end-organ dysfunction.16
Neurologic manifestations associated with enterovirus 71 infection include aseptic meningitis, a poliomyelitis-like syndrome, brainstem encephalitis, neurogenic pulmonary edema, opsoclonus-myoclonus syndrome, cerebellar ataxia, Guillain-Barré syndrome, and transverse myelitis.
Because some patients who have neurologic disease respond to treatment with high-dose methylprednisolone and intravenous immune globulin, there is reason to suspect that an autoimmune phenomenon triggered by the culprit enterovirus may be the cause of many of the neurologic symptoms.25
A 2012 meta-analysis26 found that an elevated white blood cell count and hyperglycemia could be clinically useful in distinguishing benign from severe HFMD. In patients with benign HFMD, white blood cell counts and blood glucose values were no different from those in healthy controls.26
DIAGNOSIS IS USUALLY CLINICAL
Most enteroviral infections are asymptomatic, but HFMD is a possibility if a patient has mild illness, fever, and a maculopapular or vesicular rash on the palms of the hands and soles of the feet, sometimes associated with oral ulcers (herpangina). Skin lesions can also be found on the legs, face, buttocks, and trunk.
In the United States, HFMD most commonly occurs in children under age 4 and is usually caused by coxsackievirus A16. Adults can also be affected, especially if they were in contact with children in child care, which was the case in approximately half of nonpediatric patients who tested positive for HFMD during an outbreak in several states between November 2011 and February 2012.3
The clinical characteristics of HFMD caused by enterovirus 71 may be somewhat different, with smaller vesicles, diffuse erythema of the trunk and limbs, and higher fever (temperature ≥ 39°C [102.2°F] for more than 3 days).27 However, the rash of coxsackievirus A16 HFMD may be more extensive and severe.
Other clinical manifestations of HFMD include nail dystrophies such as Beau lines and nail shedding, hyperglycemia, dehydration, and more serious and potentially life-threatening complications such as pulmonary edema28 and viral meningoencephalitis.29
Laboratory testing
In mild cases of HFMD, particularly in patients with a high probability of having the disease based on their clinical characteristics and sick contacts, laboratory testing is not necessary. Testing is usually reserved for severe cases and public health investigation of outbreaks.
Viral culture is the gold standard for diagnosing HFMD, but the final results can take nearly a week.
Polymerase chain reaction testing is faster, with a turnaround time of less than 1 day. It identifies viral RNA and is highly sensitive for detecting central nervous system infection.30
Where should samples be collected? Serum viremia precedes invasion of the skin and mucous membranes, so plasma can be tested. Inside the body, enteroviruses initially replicate in the gastrointestinal tract, although collecting a rectal swab or a stool sample is somewhat invasive. Further, in an enterovirus 71 epidemic in Taiwan, 93% of the patients had positive throat swabs, but only 30% tested positive by rectal swabs or analysis of the feces.27 At present, throat and vesicle specimens are considered to be the most useful sources for diagnostic purposes.16
ELISAs. Newly developed IgM-capture enzyme-linked immunosorbent assays (ELISAs) for coxsackievirus A16 and enterovirus 71 appear quite promising for diagnosing HFMD. These tests are inexpensive and detect IgM antibodies early and in a high percentage of patients. In the first week of the disease, the IgM detection rate was found to be 90.2% for enterovirus 71 and 68% for coxsackievirus A16.31
Cross-reactivity between these two viruses was a problem with ELISA testing in the past, causing false-positive results for enterovirus 71 in patients who in fact had coxsackievirus A16. The problem appears to be resolved in new versions that use specific enterovirus 71 proteins, eg, VP1.32
RECOGNITION AND PREVENTION ARE THE BEST MEDICINE
Recognizing HFMD early is crucial, because making the clinical diagnosis can identify patients who have signs of severe disease and can help protect future contacts and decrease the risk of an epidemic.
Infected patients continue to shed the virus for a long time, making hand hygiene and environmental control measures in health care settings and daycare centers of vital importance, to prevent spread of the infection.
Enteroviruses are stable in the environment and therefore capable of fecal-oral and oral-oral transmission. Humans are the only known natural hosts. No chemoprophylaxis or vaccination has been established to prevent HFMD. The recurrence of large-scale epidemics in the developing world is perhaps explained by ineffective sewage treatment and limited access to clean drinking water, especially in light of the fecal-oral spread of the virus. Intrafamilial spread of HFMD has been shown to be an important means of disease transmission, and asymptomatic adult carriers of these viruses may spread it to young children.33
The different viruses that cause HFMD result in a similar clinical presentation in most patients. Therefore, identifying HFMD caused by enterovirus 71, which carries a risk of severe and even fatal disease in young children vs a virus such as coxsackievirus A16, can be very difficult in practice without virologic testing. Thus, when diagnosed with HFMD, patients should be counseled to control all variables that could lead to further spread of the disease.
An analysis of epidemics in Asia suggested that public health awareness may have averted deaths in successive epidemics, highlighting the need to identify HFMD epidemics in communities and to educate patients and families about measures to prevent further spread of the virus in addition to standard supportive care.34
The CDC recommends35:
- Frequent hand-washing after toileting and changing diapers
- Disinfecting frequently used surfaces and objects, including toys
- Avoiding close contact with infected individuals and sharing of personal items such as utensils and cups.
These measures should be recommended to all affected patients.35
NO PROVEN ANTIVIRAL TREATMENT
No proven antiviral treatment exists for HFMD. Thus, the goals of treatment are typically supportive, as for any self-limited viral syndrome.16
Does acyclovir help? Shelley et al36 treated 13 patients (12 children and 1 adult) with acyclovir within 1 to 2 days of the onset of the HFMD rash and reported that it was beneficial, with significant relief of fever and skin lesions within 24 hours of starting therapy. These anecdotal results have not been replicated, and acyclovir is not an established treatment for HFMD.
If acyclovir does help, how does it work? Acyclovir, like other common antiviral medications, inactivates thymidine kinase, an enzyme produced by herpesviruses but not by HFMD-causing viruses like coxsackievirus A16. Shelley et al proposed that acyclovir may enhance the antiviral effect of the patient’s own interferon.36
Intravenous immunoglobulin has been used in severe cases during outbreaks in Asia, with retrospective data showing a potential ability to halt disease progression if used before the development of cardiopulmonary failure. However, this has not been studied prospectively and is not currently recommended.16
Acknowledgment: We would like to thank Dr. Salvador Alvarez of the Mayo Clinic Department of Infectious Disease and Dr. Donald Lookingbill of the Mayo Clinic Department of Dermatology for their collaboration.
Hand, foot, and mouth disease (HFMD) is typically a benign childhood infection—except when it isn’t so benign or when it occurs in adults.
The usual presentation is in a child with fever, oral ulcerations, and papules on the palms of the hands and the soles of the feet.1 However, severe complications can occur, including central nervous system involvement and cardiopulmonary failure, and can lead to significant morbidity and even death.2 Fortunately, these complications are rare.
Less common in North America than in other regions, HFMD has recurrently broken out in many areas of Southern Asia and the surrounding Pacific region. However, several North American outbreaks have been documented in recent years and have affected unexpected numbers of immunocompetent adults, demonstrating that this disease is of worldwide importance in adults as well as children.3
Because HFMD has the potential to reach epidemic levels in the United States, early recognition is paramount, and primary care physicians need to be familiar with its common signs and symptoms.
USUALLY A SUMMER DISEASE
HFMD occurs all around the world, exhibiting seasonal variation in temperate climates. In these locations, individual cases and regional outbreaks usually occur in the spring, summer, and fall. No sexual predisposition has been documented. Most symptomatic cases are in children under the age of 10.
OUTBREAKS AROUND THE WORLD
The disease was first described more than 40 years ago, with several large outbreaks in the last 16 years.
1998—An outbreak in Taiwan affected more than 1.5 million people, mostly children. Severe cases numbered just over 400, and 78 children died.4
2008—China,5 Singapore,6 Vietnam,7 Mongolia,8 and Brunei9 were stricken with an outbreak that affected 30,000 people and led to more than 50 deaths.
2009—An outbreak in the Henan and Shandong provinces of eastern China killed 35 people.10
2010—In several southern Chinese regions, more than 70,000 people were infected, with almost 600 deaths.11
2011 to the present. The United States has had several outbreaks in the last 3 years. Although HFMD is not one of the diseases that must be reported to public health authorities in the United States, from November 2011 to February 2012 the US Centers for Disease Control and Prevention (CDC) received reports of 63 possible cases: 38 in Alabama, 17 in Nevada, 7 in California, and 1 in Connecticut.1 Fifteen of the patients were adults, and more than half had contacts who were sick.
The most recent US outbreak, in Alabama,12 was atypical because it occurred in the winter.
CAUSED BY ENTEROVIRUSES
HFMD is caused by infection with a variety of viruses in the genus Enterovirus, a large group that in turn is part of the larger Picornaviridae family.13 The taxonomy of this genus is complicated and subject to revision; species include coxsackieviruses, polioviruses, enteroviruses, and echoviruses. They are all small, nonenveloped, single-stranded RNA viruses.
The most common strains that cause HFMD are coxsackievirus A16 and enterovirus 71. In addition, coxsackievirus A6 may be emerging, and many other coxsackievirus strains have been directly implicated, including A5, A7, A9, A10, B2, and B5.
Coxsackievirus A16 is the leading cause of HFMD.
Enterovirus 71 is the second most common cause of HFMD and has also caused outbreaks. It usually results in benign disease. However, among the causes of HFMD, it is associated with more prominent central nervous system involvement14 and is the most common cause of viral meningoencephalitis in children.
Coxsackievirus A6. In December 2011, the California Department of Public Health isolated a strain of coxsackievirus A6 that caused extensive rash and nail shedding.15 Among the 63 possible cases of HFMD reported to the CDC from November 2011 to February 2012, specimens for clinical testing were obtained in 34, and 25 of those demonstrated coxsackievirus A6 infection.3
FEVER, ORAL ULCERS, RASH ON HANDS AND FEET
The typical clinical manifestations of HFMD are fever, stomatitis with oral ulcers, and an exanthem affecting the palms, soles, and other parts of the body. These last less than 7 to 10 days, usually occur during the spring to fall months, and have a benign course.
The incubation period is 3 to 5 days, with a prodrome that may include fever, malaise, abdominal pain, and myalgia before the onset of oral and cutaneous findings. Painful oral ulcers may precede the exanthem and can result in dehydration.16
The cutaneous manifestation of HFMD is typically a papulovesicular rash affecting the palms, soles, and buttocks (Figure 1). Other sites may include the knees, elbows, and the dorsal surfaces of the hands and feet. The lesions may be maculopapular and can be either asymptomatic or tender and painful. Desquamation can follow the exanthem, and lesions usually resolve without scarring or secondary infection.16,17

Table 1 and Table 2 compare HFMD with other common illnesses that can cause similar skin and mucosal findings. In particular, herpangina has the identical clinical presentation as HFMD except that it does not cause skin lesions. It is caused by many of the same enteroviruses linked to HFMD.
Different viruses, different signs?
The numerous viruses that cause HFMD usually cause similar signs and symptoms during bouts of typical, self-limited disease. However, neurologic and cardiopulmonary involvement, which are fortunately rare, are more often associated with enterovirus 71 infection.
Nail manifestations are common in HFMD. Nail separation from the nail matrix (onychomadesis) was associated with coxsackievirus A6 infection during a 2010 outbreak of HFMD in Taiwan and in a 2009 outbreak in Finland.18 Moreover, this virus was cultured from a nail specimen in one patient, suggesting viral infiltration as the cause of nail-matrix arrest.19
Perioral skin eruptions, desquamation, and Beau lines have also been associated with coxsackievirus A6.18 Beau lines are transverse depressions of the nail, most evident in the central nail plate; when seen on multiple nails, they imply a systemic illness causing disruption of nail matrix growth.20
Atypical HFMD and coxsackievirus A6
Atypical HFMD has recently been described in connection with coxsackievirus A6. Lott et al21 reported five cases of coxsackievirus A6-associated HFMD in 2013. Atypically, three of the affected patients presented in winter months, two were adults, and two had widespread skin involvement.21
Mathes et al22 reported a series of 80 cases of enteroviral infections in which the lesions had a predilection for the antecubital and popliteal fossae and were similar in severity and distribution to those seen in eczema herpeticum or Kaposi varicelliform eruption in patients with and patients without a history of atopic dermatitis. They named this find-clinical finding of pronounced coxsackievirus-associated skin disease at sites previously affected by atopic dermatitis.
Additional cutaneous findings of coxsackievirus A6 infection may include onychomadesis, Beau lines, and vesiculobullous lesions. Patients with atypical, coxsackievirus A6-associated HFMD may not have oral lesions.23
In the five cases reported by Lott et al,21 significant systemic symptoms (fever, chills, diarrhea, and myalgias) led all but one of the patients to seek care in an emergency department. However, atypical HFMD has not been associated with life-threatening illness.
Atypical HFMD associated with coxsackievirus A6 is an emerging entity in the United States, and the acuity of both cutaneous and systemic symptoms poses a diagnostic dilemma. Furthermore, infection has been documented in immunocompetent adults.23 Familiarity with the clinical findings may expedite appropriate care, prevent spread to contacts, and avoid unnecessary testing.
Neurologic and cardiopulmonary manifestations
Enteroviruses are the most common causes of viral meningoencephalitis in the United States. They mainly affect children and cause serious and potentially chronic disease in those with humoral immunodeficiencies.24 Neurologic and cardiopulmonary manifestations of HFMD are varied and extremely rare in the United States but should always be viewed clinically as signs of concern and severe disease.
Signs of potentially fatal disease that have been observed in young children include tachycardia, tachypnea, hypotension, hypertension, gastrointestinal bleeding, neurologic symptoms, leukocytosis, absence of oral lesions, and vomiting.2 Signs of dysautonomia, myoclonus, ataxia, and brainstem involvement may portend fatal disease in which rapid decompensation is the result of cardiogenic shock due to loss of ventricular contractility, causing pulmonary edema and end-organ dysfunction.16
Neurologic manifestations associated with enterovirus 71 infection include aseptic meningitis, a poliomyelitis-like syndrome, brainstem encephalitis, neurogenic pulmonary edema, opsoclonus-myoclonus syndrome, cerebellar ataxia, Guillain-Barré syndrome, and transverse myelitis.
Because some patients who have neurologic disease respond to treatment with high-dose methylprednisolone and intravenous immune globulin, there is reason to suspect that an autoimmune phenomenon triggered by the culprit enterovirus may be the cause of many of the neurologic symptoms.25
A 2012 meta-analysis26 found that an elevated white blood cell count and hyperglycemia could be clinically useful in distinguishing benign from severe HFMD. In patients with benign HFMD, white blood cell counts and blood glucose values were no different from those in healthy controls.26
DIAGNOSIS IS USUALLY CLINICAL
Most enteroviral infections are asymptomatic, but HFMD is a possibility if a patient has mild illness, fever, and a maculopapular or vesicular rash on the palms of the hands and soles of the feet, sometimes associated with oral ulcers (herpangina). Skin lesions can also be found on the legs, face, buttocks, and trunk.
In the United States, HFMD most commonly occurs in children under age 4 and is usually caused by coxsackievirus A16. Adults can also be affected, especially if they were in contact with children in child care, which was the case in approximately half of nonpediatric patients who tested positive for HFMD during an outbreak in several states between November 2011 and February 2012.3
The clinical characteristics of HFMD caused by enterovirus 71 may be somewhat different, with smaller vesicles, diffuse erythema of the trunk and limbs, and higher fever (temperature ≥ 39°C [102.2°F] for more than 3 days).27 However, the rash of coxsackievirus A16 HFMD may be more extensive and severe.
Other clinical manifestations of HFMD include nail dystrophies such as Beau lines and nail shedding, hyperglycemia, dehydration, and more serious and potentially life-threatening complications such as pulmonary edema28 and viral meningoencephalitis.29
Laboratory testing
In mild cases of HFMD, particularly in patients with a high probability of having the disease based on their clinical characteristics and sick contacts, laboratory testing is not necessary. Testing is usually reserved for severe cases and public health investigation of outbreaks.
Viral culture is the gold standard for diagnosing HFMD, but the final results can take nearly a week.
Polymerase chain reaction testing is faster, with a turnaround time of less than 1 day. It identifies viral RNA and is highly sensitive for detecting central nervous system infection.30
Where should samples be collected? Serum viremia precedes invasion of the skin and mucous membranes, so plasma can be tested. Inside the body, enteroviruses initially replicate in the gastrointestinal tract, although collecting a rectal swab or a stool sample is somewhat invasive. Further, in an enterovirus 71 epidemic in Taiwan, 93% of the patients had positive throat swabs, but only 30% tested positive by rectal swabs or analysis of the feces.27 At present, throat and vesicle specimens are considered to be the most useful sources for diagnostic purposes.16
ELISAs. Newly developed IgM-capture enzyme-linked immunosorbent assays (ELISAs) for coxsackievirus A16 and enterovirus 71 appear quite promising for diagnosing HFMD. These tests are inexpensive and detect IgM antibodies early and in a high percentage of patients. In the first week of the disease, the IgM detection rate was found to be 90.2% for enterovirus 71 and 68% for coxsackievirus A16.31
Cross-reactivity between these two viruses was a problem with ELISA testing in the past, causing false-positive results for enterovirus 71 in patients who in fact had coxsackievirus A16. The problem appears to be resolved in new versions that use specific enterovirus 71 proteins, eg, VP1.32
RECOGNITION AND PREVENTION ARE THE BEST MEDICINE
Recognizing HFMD early is crucial, because making the clinical diagnosis can identify patients who have signs of severe disease and can help protect future contacts and decrease the risk of an epidemic.
Infected patients continue to shed the virus for a long time, making hand hygiene and environmental control measures in health care settings and daycare centers of vital importance, to prevent spread of the infection.
Enteroviruses are stable in the environment and therefore capable of fecal-oral and oral-oral transmission. Humans are the only known natural hosts. No chemoprophylaxis or vaccination has been established to prevent HFMD. The recurrence of large-scale epidemics in the developing world is perhaps explained by ineffective sewage treatment and limited access to clean drinking water, especially in light of the fecal-oral spread of the virus. Intrafamilial spread of HFMD has been shown to be an important means of disease transmission, and asymptomatic adult carriers of these viruses may spread it to young children.33
The different viruses that cause HFMD result in a similar clinical presentation in most patients. Therefore, identifying HFMD caused by enterovirus 71, which carries a risk of severe and even fatal disease in young children vs a virus such as coxsackievirus A16, can be very difficult in practice without virologic testing. Thus, when diagnosed with HFMD, patients should be counseled to control all variables that could lead to further spread of the disease.
An analysis of epidemics in Asia suggested that public health awareness may have averted deaths in successive epidemics, highlighting the need to identify HFMD epidemics in communities and to educate patients and families about measures to prevent further spread of the virus in addition to standard supportive care.34
The CDC recommends35:
- Frequent hand-washing after toileting and changing diapers
- Disinfecting frequently used surfaces and objects, including toys
- Avoiding close contact with infected individuals and sharing of personal items such as utensils and cups.
These measures should be recommended to all affected patients.35
NO PROVEN ANTIVIRAL TREATMENT
No proven antiviral treatment exists for HFMD. Thus, the goals of treatment are typically supportive, as for any self-limited viral syndrome.16
Does acyclovir help? Shelley et al36 treated 13 patients (12 children and 1 adult) with acyclovir within 1 to 2 days of the onset of the HFMD rash and reported that it was beneficial, with significant relief of fever and skin lesions within 24 hours of starting therapy. These anecdotal results have not been replicated, and acyclovir is not an established treatment for HFMD.
If acyclovir does help, how does it work? Acyclovir, like other common antiviral medications, inactivates thymidine kinase, an enzyme produced by herpesviruses but not by HFMD-causing viruses like coxsackievirus A16. Shelley et al proposed that acyclovir may enhance the antiviral effect of the patient’s own interferon.36
Intravenous immunoglobulin has been used in severe cases during outbreaks in Asia, with retrospective data showing a potential ability to halt disease progression if used before the development of cardiopulmonary failure. However, this has not been studied prospectively and is not currently recommended.16
Acknowledgment: We would like to thank Dr. Salvador Alvarez of the Mayo Clinic Department of Infectious Disease and Dr. Donald Lookingbill of the Mayo Clinic Department of Dermatology for their collaboration.
- Centers for Disease Control and Prevention (CDC). Hand, Foot, and Mouth Disease (HFMD). www.cdc.gov/hand-foot-mouth/index.html. Accessed June 10, 2014.
- Chatproedprai S, Theanboonlers A, Korkong S, Thongmee C, Wananukul S, Poovorawan Y. Clinical and molecular characterization of hand-foot-and-mouth disease in Thailand, 2008–2009. Jpn J Infect Dis 2010; 63:229–233.
- Centers for Disease Control and Prevention (CDC). Notes from the field: severe hand, foot, and mouth disease associated with coxsackievirus A6—Alabama, Connecticut, California, and Nevada, November 2011–February 2012. MMWR Morb Mortal Wkly Rep 2012; 61:213–214.
- Ho M, Chen ER, Hsu KH, et al. An epidemic of enterovirus 71 infection in Taiwan. Taiwan Enterovirus Epidemic Working Group. N Engl J Med 1999; 341:929–935.
- BBC News. China virus toll continues rise. May 5, 2008. http://news.bbc.co.uk/2/hi/asia-pacific/7383796.stm. Accessed February 5, 2014.
- Suhaimi ND. HFMD: 1,000 cases a week in Singapore is unusual, says doc. Straits Times April 20, 2008.
- Viet Nam News: HFMD cases prompt tighter health screening at airport. May 15, 2008.
- UBPOST. EV-71 virus continues dramatic rise. May 22, 2008.
- Begawan BS. 1,053 HFMD cases recorded. Brunei Times. November 7, 2008.
- Chinaview. Hand-foot-mouth disease death toll rises to 17 in East China’s Shandong Province. April 9, 2009.
- Chinaview. China reports 537 deaths from hand-foot-mouth disease this year. June 24, 2010.
- Wolfson H. Outbreak of hand, foot and mouth disease severe in Alabama. Birmingham News February 13, 2012.
- Centers for Disease Control and Prevention (CDC). Non-Polio Enterovirus Infections. www.cdc.gov/non-polio-enterovirus/. Accessed June 10, 2014.
- Chan KP, Goh KT, Chong CY, Teo ES, Lau G, Ling AE. Epidemic hand, foot and mouth disease caused by human enterovirus 71, Singapore. Emerg Infect Dis 2003; 9:78–85.
- California Department of Public Health. Coxsackievirus A6 (CVA6). 2011. www.cdph.ca.gov/programs/cder/Pages/CVA6.aspx. Accessed June 10, 2014.
- World Health Organization: Western Pacific Region. A Guide to Clinical management and Public Health Response for Hand, Foot, and Mouth Disease (HFMD).
- Shin JU, Oh SH, Lee JH. A case of hand-foot-mouth disease in an immunocompetent adult. Ann Dermatol 2010; 22:216–218.
- Wei SH, Huang YP, Liu MC, et al. An outbreak of coxsackievirus A6 hand, foot, and mouth disease associated with onychomadesis in Taiwan, 2010. BMC Infect Dis 2011; 11:346.
- Osterback R, Vuorinen T, Linna M, Susi P, Hyypiä T, Waris M. Coxsackievirus A6 and hand, foot, and mouth disease, Finland. Emerg Infect Dis 2009; 15:1485–1488.
- Tosti A, Piraccini BM. Nail Disorders. In:Bolognia JL, Jorizzo JL, Schaffer JV, editors. Dermatology JV. 3rded. Elsevier Limited; 2012:1129–1144.
- Lott JP, Liu K, Landry ML, et al. Atypical hand-foot-and-mouth disease associated with coxsackievirus A6 infection. J Am Acad Dermatol 2013; 69:736–741.
- Mathes EF, Oza V, Frieden IJ, et al. ”Eczema coxsackium” and unusual cutaneous findings in an enterovirus outbreak. Pediatrics 2013; 132:e149–e157.
- Kaminska K, Martinetti G, Lucchini R, Kaya G, Mainetti C. Coxsackievirus A6 and hand, foot and mouth disease: three case reports of familial child-to-immunocompetent adult transmission and a literature review. Case Rep Dermatol 2013; 5:203–209.
- Romero JR. Diagnosis and management of enteroviral infections of the central nervous system. Curr Infect Dis Rep 2002; 4:309–316.
- Akiyama K, Imazeki R, Yoshii F, Koide T, Muto J. An adult case of hand, foot, and mouth disease caused by enterovirus 71 accompanied by opsoclonus myoclonica. Tokai J Exp Clin Med 2008; 33:143–145.
- Li Y, Zhu R, Qian Y, Deng J. The characteristics of blood glucose and WBC counts in peripheral blood of cases of hand foot and mouth disease in China: a systematic review. PLoS One 2012; 7:e29003.
- Chang LY, King CC, Hsu KH, et al. Risk factors of enterovirus 71 infection and associated hand, foot, and mouth disease/herpangina in children during an epidemic in Taiwan. Pediatrics 2002; 109:e88.
- Wang SM, Liu CC, Tseng HW, et al. Clinical spectrum of enterovirus 71 infection in children in southern Taiwan, with an emphasis on neurological complications. Clin Infect Dis 1999; 29:184–190.
- Chang LY, Lin TY, Hsu KH, et al. Clinical features and risk factors of pulmonary oedema after enterovirus-71-related hand, foot, and mouth disease. Lancet 1999; 354:1682–1686.
- Mayo Clinic Laboratories. Enterovirus, Molecular Detection, PCR, Plasma. www.mayomedicallaboratories.com/test-catalog/Clinical+and+Interpretive/89893. Accessed June 10, 2014.
- Yu N, Guo M, He SJ, et al. Evaluation of human enterovirus 71 and coxsackievirus A16 specific immunoglobulin M antibodies for diagnosis of hand-foot-and-mouth disease. Virol J 2012; 9:12.
- Wang C, You A, Tian X, et al. Analysis and solution of false-positives when testing CVA16 sera using an antibody assay against the EV71 virus. Virus Res 2013; 176:33–36.
- Liu MY, Liu W, Luo J, et al. Characterization of an outbreak of hand, foot, and mouth disease in Nanchang, China in 2010. PLoS One 2011; 6:e25287.
- Zhang J, Sun J, Chang Z, Zhang W, Wang Z, Feng Z. Characterization of hand, foot, and mouth disease in China between 2008 and 2009. Biomed Environ Sci 2011; 24:214–221.
- Centers for Disease Control and Prevention (CDC). Hand, Foot, and Mouth Disease: Prevention & Treatment. www.cdc.gov/hand-foot-mouth/about/prevention-treatment.html. Accessed June 10, 2014.
- Shelley WB, Hashim M, Shelley ED. Acyclovir in the treatment of hand-foot-and-mouth disease. Cutis 1996; 57:232–234.
- Centers for Disease Control and Prevention (CDC). Hand, Foot, and Mouth Disease (HFMD). www.cdc.gov/hand-foot-mouth/index.html. Accessed June 10, 2014.
- Chatproedprai S, Theanboonlers A, Korkong S, Thongmee C, Wananukul S, Poovorawan Y. Clinical and molecular characterization of hand-foot-and-mouth disease in Thailand, 2008–2009. Jpn J Infect Dis 2010; 63:229–233.
- Centers for Disease Control and Prevention (CDC). Notes from the field: severe hand, foot, and mouth disease associated with coxsackievirus A6—Alabama, Connecticut, California, and Nevada, November 2011–February 2012. MMWR Morb Mortal Wkly Rep 2012; 61:213–214.
- Ho M, Chen ER, Hsu KH, et al. An epidemic of enterovirus 71 infection in Taiwan. Taiwan Enterovirus Epidemic Working Group. N Engl J Med 1999; 341:929–935.
- BBC News. China virus toll continues rise. May 5, 2008. http://news.bbc.co.uk/2/hi/asia-pacific/7383796.stm. Accessed February 5, 2014.
- Suhaimi ND. HFMD: 1,000 cases a week in Singapore is unusual, says doc. Straits Times April 20, 2008.
- Viet Nam News: HFMD cases prompt tighter health screening at airport. May 15, 2008.
- UBPOST. EV-71 virus continues dramatic rise. May 22, 2008.
- Begawan BS. 1,053 HFMD cases recorded. Brunei Times. November 7, 2008.
- Chinaview. Hand-foot-mouth disease death toll rises to 17 in East China’s Shandong Province. April 9, 2009.
- Chinaview. China reports 537 deaths from hand-foot-mouth disease this year. June 24, 2010.
- Wolfson H. Outbreak of hand, foot and mouth disease severe in Alabama. Birmingham News February 13, 2012.
- Centers for Disease Control and Prevention (CDC). Non-Polio Enterovirus Infections. www.cdc.gov/non-polio-enterovirus/. Accessed June 10, 2014.
- Chan KP, Goh KT, Chong CY, Teo ES, Lau G, Ling AE. Epidemic hand, foot and mouth disease caused by human enterovirus 71, Singapore. Emerg Infect Dis 2003; 9:78–85.
- California Department of Public Health. Coxsackievirus A6 (CVA6). 2011. www.cdph.ca.gov/programs/cder/Pages/CVA6.aspx. Accessed June 10, 2014.
- World Health Organization: Western Pacific Region. A Guide to Clinical management and Public Health Response for Hand, Foot, and Mouth Disease (HFMD).
- Shin JU, Oh SH, Lee JH. A case of hand-foot-mouth disease in an immunocompetent adult. Ann Dermatol 2010; 22:216–218.
- Wei SH, Huang YP, Liu MC, et al. An outbreak of coxsackievirus A6 hand, foot, and mouth disease associated with onychomadesis in Taiwan, 2010. BMC Infect Dis 2011; 11:346.
- Osterback R, Vuorinen T, Linna M, Susi P, Hyypiä T, Waris M. Coxsackievirus A6 and hand, foot, and mouth disease, Finland. Emerg Infect Dis 2009; 15:1485–1488.
- Tosti A, Piraccini BM. Nail Disorders. In:Bolognia JL, Jorizzo JL, Schaffer JV, editors. Dermatology JV. 3rded. Elsevier Limited; 2012:1129–1144.
- Lott JP, Liu K, Landry ML, et al. Atypical hand-foot-and-mouth disease associated with coxsackievirus A6 infection. J Am Acad Dermatol 2013; 69:736–741.
- Mathes EF, Oza V, Frieden IJ, et al. ”Eczema coxsackium” and unusual cutaneous findings in an enterovirus outbreak. Pediatrics 2013; 132:e149–e157.
- Kaminska K, Martinetti G, Lucchini R, Kaya G, Mainetti C. Coxsackievirus A6 and hand, foot and mouth disease: three case reports of familial child-to-immunocompetent adult transmission and a literature review. Case Rep Dermatol 2013; 5:203–209.
- Romero JR. Diagnosis and management of enteroviral infections of the central nervous system. Curr Infect Dis Rep 2002; 4:309–316.
- Akiyama K, Imazeki R, Yoshii F, Koide T, Muto J. An adult case of hand, foot, and mouth disease caused by enterovirus 71 accompanied by opsoclonus myoclonica. Tokai J Exp Clin Med 2008; 33:143–145.
- Li Y, Zhu R, Qian Y, Deng J. The characteristics of blood glucose and WBC counts in peripheral blood of cases of hand foot and mouth disease in China: a systematic review. PLoS One 2012; 7:e29003.
- Chang LY, King CC, Hsu KH, et al. Risk factors of enterovirus 71 infection and associated hand, foot, and mouth disease/herpangina in children during an epidemic in Taiwan. Pediatrics 2002; 109:e88.
- Wang SM, Liu CC, Tseng HW, et al. Clinical spectrum of enterovirus 71 infection in children in southern Taiwan, with an emphasis on neurological complications. Clin Infect Dis 1999; 29:184–190.
- Chang LY, Lin TY, Hsu KH, et al. Clinical features and risk factors of pulmonary oedema after enterovirus-71-related hand, foot, and mouth disease. Lancet 1999; 354:1682–1686.
- Mayo Clinic Laboratories. Enterovirus, Molecular Detection, PCR, Plasma. www.mayomedicallaboratories.com/test-catalog/Clinical+and+Interpretive/89893. Accessed June 10, 2014.
- Yu N, Guo M, He SJ, et al. Evaluation of human enterovirus 71 and coxsackievirus A16 specific immunoglobulin M antibodies for diagnosis of hand-foot-and-mouth disease. Virol J 2012; 9:12.
- Wang C, You A, Tian X, et al. Analysis and solution of false-positives when testing CVA16 sera using an antibody assay against the EV71 virus. Virus Res 2013; 176:33–36.
- Liu MY, Liu W, Luo J, et al. Characterization of an outbreak of hand, foot, and mouth disease in Nanchang, China in 2010. PLoS One 2011; 6:e25287.
- Zhang J, Sun J, Chang Z, Zhang W, Wang Z, Feng Z. Characterization of hand, foot, and mouth disease in China between 2008 and 2009. Biomed Environ Sci 2011; 24:214–221.
- Centers for Disease Control and Prevention (CDC). Hand, Foot, and Mouth Disease: Prevention & Treatment. www.cdc.gov/hand-foot-mouth/about/prevention-treatment.html. Accessed June 10, 2014.
- Shelley WB, Hashim M, Shelley ED. Acyclovir in the treatment of hand-foot-and-mouth disease. Cutis 1996; 57:232–234.
KEY POINTS
- In Asian and Pacific nations, HFMD has been a significant public health concern since 1997, with recurrent epidemics and, in some cases, severe complications, including central nervous system disease, pulmonary edema, and death.
- Coxsackievirus A16 and enterovirus 71 are the most common agents of HFMD. In addition, coxsackievirus A6 seems to be emerging.
- Neurologic and cardiopulmonary involvement are more often associated with enterovirus 71 infection.
- In March 2012, 63 cases of severe HFMD were reported in Alabama, California, Connecticut, and Nevada. Fifteen of the patients were adults, and more than half had positive sick contacts. Of the 34 patients who underwent serologic testing, 25 were positive for coxsackievirus A6, an unusual pathogen for HFMD in the United States, associated with more severe skin findings.
- Treatment focuses on supportive care and prevention.
Can we reduce the risk of readmission for a patient with an exacerbation of COPD?
We think so. Some strategies to reduce readmission rates, such as coordinating care and managing comorbidities, apply to chronic diseases in general, while others are disease-specific. To reduce the need for hospital readmission for chronic obstructive pulmonary disease (COPD), coordinated efforts involving both inpatient and outpatient care are necessary. This can be achieved by using a checklist before discharge (Table 1) and by implementing outpatient COPD programs that continue patient education and provide rapid access to medical support if needed.
There is room for improvement. COPD is common and expensive, with high rates of hospital readmission,1 and up to 70% of the money we spend on it goes for hospital care.2 No wonder then that the Centers for Medicare and Medicaid Services has now expanded its Readmissions Reduction Program to include acute COPD exacerbations.3 Yet in a retrospective study, Yip et al4 found that fewer than half of patients hospitalized with acute exacerbation of COPD received appropriate vaccinations, counseling on smoking cessation, and long-acting inhalers—all of which are on our checklist.4
The following interventions have been demonstrated to be useful in reducing COPD hospital admissions and the risk of death.
SMOKING CESSATION
Cigarette smoking is the most common and easily identifiable risk factor for COPD exacerbation.5
Au et al5 found that quitting smoking reduces the risk of COPD exacerbation (adjusted hazard ratio 0.78, 95% confidence interval [CI] 0.75–0.87), and the risk keeps decreasing the longer the patient stays off tobacco.5
Whether counseling hospitalized patients on smoking cessation reduces the COPD readmission rate has not been well studied. However, a meta-analysis of nine randomized controlled trials, two of which were done in the hospital, revealed higher abstinence rates in COPD patients who received extensive counseling on smoking cessation.7 For these reasons, hospitalized COPD patients who smoke should be strongly encouraged to quit.6
PNEUMOCOCCAL AND INFLUENZA VACCINATIONS
In a large retrospective study,8 pneumococcal vaccination was associated with a significantly lower risk of hospitalization for pneumonia in patients with chronic lung disease, including those with COPD (relative risk [RR] 0.57, 95% CI 0.38–0.84). The benefit was even greater with pneumococcal and influenza vaccinations during the influenza season (RR 0.28, 95% CI 0.14–0.58).
Randomized controlled trials indicate that influenza vaccination may reduce the rate of COPD exacerbations, especially in epidemic years when the proportion of exacerbations due to influenza is higher.9
Wongsurakiat et al10 found a significant reduction in the incidence of influenza-related acute respiratory illness in COPD patients in a well-designed randomized, placebo-controlled trial (RR 0.24, P = .005).10
Similarly, in another randomized controlled trial, pneumococcal vaccination was effective in preventing community-acquired pneumonia in COPD patients under age 65 and in those with severe airflow obstruction, although no statistically significant differences were found among other groups of patients with COPD.11
Therefore, influenza and pneumococcal vaccinations are recommended by major COPD guidelines, such as GOLD (Global Strategy for the Diagnosis, Management, and Prevention of Chronic Obstructive Lung Disease).6
INHALERS
Inhaler therapy is recommended based on COPD severity according to GOLD classification, and appropriate inhaler therapy with proper inhaler technique reduces the number of COPD exacerbations and hospitalizations.6
Long-acting beta-agonists and anticholinergics reduce the risk of COPD exacerbation and hospitalization and so are preferred over short-acting formulations except for patients in GOLD group A, ie, those who have few symptoms and are at low risk of exacerbations.6
Long-term treatment with inhaled corticosteroids with long-acting bronchodilators is recommended for patients at high risk of exacerbations (ie, those with two or more exacerbations in the previous year or a forced expiratory volume in 1 second [FEV1] less than 50% of predicted).6
OXYGEN THERAPY
Two older randomized controlled trials, the Nocturnal Oxygen Therapy Trial and the Medical Research Council study, reviewed by Stoller et al,12 provided clear evidence that oxygen therapy reduces the death rate and improves quality of life in COPD patients who have chronic resting hypoxemia (room air Pao2 ≤ 55 mm Hg, or ≤ 59 mm Hg with signs of right-sided heart strain or polycythemia).
PULMONARY REHABILITATION
Pulmonary rehabilitation likely reduces hospital admissions by improving exercise capacity.13 A systematic review of six trials in 230 patients found that respiratory rehabilitation after an acute COPD exacerbation reduced the risk of COPD hospital admission (RR 0.26, 95% CI 0.12–0.54) and the risk of death (RR 0.45, 95% CI 0.22–0.91).13
OTHER INTERVENTIONS
Home noninvasive ventilator support reduced hospital and intensive care unit readmissions in select patients recurrently hospitalized for acidotic exacerbations of COPD in one small study.14
Long-term antibiotic therapy. Although there is evidence that azithromycin, taken daily for 1 year, decreases the frequency of COPD exacerbations,15 concern persists that this approach promotes antibiotic resistance, and the GOLD guidelines do not recommend routinely using antibiotics in patients with clinically stable COPD.6
Roflumilast. According to the GOLD guidelines, the phosphodiesterase-4 inhibitor roflumilast (Daliresp) may be useful in reducing exacerbations in patients who have an FEV1 less than 50% of predicted, chronic bronchitis, and frequent exacerbations.6
Referral. Patients who have severe recurrent COPD exacerbations despite appropriate therapy will likely benefit from referral to a pulmonary specialist for other options such as theophylline, lung-reduction surgery, and lung transplantation.
PATIENT EDUCATION AND OUTPATIENT COPD PROGRAMS
There is growing evidence that outpatient programs that provide education and medical support significantly reduce the rate of hospitalizations for COPD.16–18 Patient education includes symptom monitoring, early recognition of an exacerbation, appropriate use of inhalers and nebulizers, and advice on smoking cessation.16
On the other hand, a Veterans Administration randomized controlled trial was stopped early because of a higher rate of death in the group that underwent a comprehensive care-management program of COPD education, an action plan for identification and treatment of exacerbations, and scheduled proactive telephone calls for case management.19
Further study is needed to investigate the cost-effectiveness and safety of COPD management programs and whether to adopt such programs on a systematic level.
In conclusion, COPD patients require a comprehensive approach based on studied interventions. This may be achieved through systematic methods that allow each patient to benefit from all possible interventions appropriate for him or her. Hospitalization of COPD patients provides an excellent opportunity to implement this comprehensive approach.
- Westert GP, Lagoe RJ, Keskimäki I, Leyland A, Murphy M. An international study of hospital readmissions and related utilization in Europe and the USA. Health Policy 2002; 61:269–278.
- Halpern MT, Stanford RH, Borker R. The burden of COPD in the USA: results from the Confronting COPD survey. Respir Med 2003; 97(suppl C):S81–S89.
- Centers for Medicare and Medicaid Services. Readmissions reduction program. www.cms.gov/Medicare/Medicare-Fee-for-Service-Payment/AcuteInpatientPPS/Readmissions-Reduction-Program.html. Accessed August 9, 2014.
- Yip NH, Yuen G, Lazar EJ, et al. Analysis of hospitalizations for COPD exacerbation: opportunities for improving care. COPD 2010; 7:85–92.
- Au DH, Bryson CL, Chien JW, et al. The effects of smoking cessation on the risk of chronic obstructive pulmonary disease exacerbations. J Gen Intern Med 2009; 24:457–463.
- Vestbo J, Hurd SS, Agustí AG, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med 2013; 187:347–365.
- Thabane MCOPD Working Group. Smoking cessation for patients with chronic obstructive pulmonary disease (COPD): an evidence-based analysis. Ont Health Technol Assess Ser 2012; 12:1–50.
- Nichol KL, Baken L, Wuorenma J, Nelson A. The health and economic benefits associated with pneumococcal vaccination of elderly persons with chronic lung disease. Arch Intern Med 1999; 159:2437–2442.
- Poole PJ, Chacko E, Wood-Baker RW, Cates CJ. Influenza vaccine for patients with chronic obstructive pulmonary disease. Cochrane Database Syst Rev 2006; 1:CD002733.
- Wongsurakiat P, Maranetra KN, Wasi C, Kositanont U, Dejsomritrutai W, Charoenratanakul S. Acute respiratory illness in patients with COPD and the effectiveness of influenza vaccination: a randomized controlled study. Chest 2004; 125:2011–2020.
- Alfageme I, Vazquez R, Reyes N, et al. Clinical efficacy of anti-pneumococcal vaccination in patients with COPD. Thorax 2006; 61:189–195.
- Stoller JK, Panos RJ, Krachman S, Doherty DE, Make B; Long-term Oxygen Treatment Trial Research Group. Oxygen therapy for patients with COPD: current evidence and the long-term oxygen treatment trial. Chest 2010; 138:179–187.
- Puhan MA, Scharplatz M, Troosters T, Steurer J. Respiratory rehabilitation after acute exacerbation of COPD may reduce risk for readmission and mortality—a systematic review. Respir Res 2005; 6:54.
- Tuggey JM, Plant PK, Elliott MW. Domiciliary non-invasive ventilation for recurrent acidotic exacerbations of COPD: an economic analysis. Thorax 2003; 58:867–871.
- Albert RK, Connett J, Bailey WC, et al; COPD Clinical Research Network. Azithromycin for prevention of exacerbations of COPD. N Engl J Med 2011; 365:689–698.
- Lawlor M, Kealy S, Agnew M, et al. Early discharge care with ongoing follow-up support may reduce hospital readmissions in COPD. Int J Chron Obstruct Pulmon Dis 2009; 4:55–60.
- Gadoury MA, Schwartzman K, Rouleau M, et al; Chronic Obstructive Pulmonary Disease axis of the Respiratory Health Network, Fonds de la Recherche en Santé du Québec (FRSQ). Self-management reduces both short- and long-term hospitalisation in COPD. Eur Respir J 2005; 26:853–857.
- Rice KL, Dewan N, Bloomfield HE, et al. Disease management program for chronic obstructive pulmonary disease: a randomized controlled trial. Am J Respir Crit Care Med 2010; 182:890–896.
- Fan VS, Gaziano JM, Lew R, et al. A comprehensive care management program to prevent chronic obstructive pulmonary disease hospitalizations: a randomized, controlled trial. Ann Intern Med 2012; 156:673–683.
- COPD Working Group. Noninvasive positive pressure ventilation for chronic respiratory failure patients with stable chronic obstructive pulmonary disease (COPD): an evidence-based analysis. Ont Health Technol Assess Ser 2012; 12( 9):1–51.
We think so. Some strategies to reduce readmission rates, such as coordinating care and managing comorbidities, apply to chronic diseases in general, while others are disease-specific. To reduce the need for hospital readmission for chronic obstructive pulmonary disease (COPD), coordinated efforts involving both inpatient and outpatient care are necessary. This can be achieved by using a checklist before discharge (Table 1) and by implementing outpatient COPD programs that continue patient education and provide rapid access to medical support if needed.
There is room for improvement. COPD is common and expensive, with high rates of hospital readmission,1 and up to 70% of the money we spend on it goes for hospital care.2 No wonder then that the Centers for Medicare and Medicaid Services has now expanded its Readmissions Reduction Program to include acute COPD exacerbations.3 Yet in a retrospective study, Yip et al4 found that fewer than half of patients hospitalized with acute exacerbation of COPD received appropriate vaccinations, counseling on smoking cessation, and long-acting inhalers—all of which are on our checklist.4
The following interventions have been demonstrated to be useful in reducing COPD hospital admissions and the risk of death.
SMOKING CESSATION
Cigarette smoking is the most common and easily identifiable risk factor for COPD exacerbation.5
Au et al5 found that quitting smoking reduces the risk of COPD exacerbation (adjusted hazard ratio 0.78, 95% confidence interval [CI] 0.75–0.87), and the risk keeps decreasing the longer the patient stays off tobacco.5
Whether counseling hospitalized patients on smoking cessation reduces the COPD readmission rate has not been well studied. However, a meta-analysis of nine randomized controlled trials, two of which were done in the hospital, revealed higher abstinence rates in COPD patients who received extensive counseling on smoking cessation.7 For these reasons, hospitalized COPD patients who smoke should be strongly encouraged to quit.6
PNEUMOCOCCAL AND INFLUENZA VACCINATIONS
In a large retrospective study,8 pneumococcal vaccination was associated with a significantly lower risk of hospitalization for pneumonia in patients with chronic lung disease, including those with COPD (relative risk [RR] 0.57, 95% CI 0.38–0.84). The benefit was even greater with pneumococcal and influenza vaccinations during the influenza season (RR 0.28, 95% CI 0.14–0.58).
Randomized controlled trials indicate that influenza vaccination may reduce the rate of COPD exacerbations, especially in epidemic years when the proportion of exacerbations due to influenza is higher.9
Wongsurakiat et al10 found a significant reduction in the incidence of influenza-related acute respiratory illness in COPD patients in a well-designed randomized, placebo-controlled trial (RR 0.24, P = .005).10
Similarly, in another randomized controlled trial, pneumococcal vaccination was effective in preventing community-acquired pneumonia in COPD patients under age 65 and in those with severe airflow obstruction, although no statistically significant differences were found among other groups of patients with COPD.11
Therefore, influenza and pneumococcal vaccinations are recommended by major COPD guidelines, such as GOLD (Global Strategy for the Diagnosis, Management, and Prevention of Chronic Obstructive Lung Disease).6
INHALERS
Inhaler therapy is recommended based on COPD severity according to GOLD classification, and appropriate inhaler therapy with proper inhaler technique reduces the number of COPD exacerbations and hospitalizations.6
Long-acting beta-agonists and anticholinergics reduce the risk of COPD exacerbation and hospitalization and so are preferred over short-acting formulations except for patients in GOLD group A, ie, those who have few symptoms and are at low risk of exacerbations.6
Long-term treatment with inhaled corticosteroids with long-acting bronchodilators is recommended for patients at high risk of exacerbations (ie, those with two or more exacerbations in the previous year or a forced expiratory volume in 1 second [FEV1] less than 50% of predicted).6
OXYGEN THERAPY
Two older randomized controlled trials, the Nocturnal Oxygen Therapy Trial and the Medical Research Council study, reviewed by Stoller et al,12 provided clear evidence that oxygen therapy reduces the death rate and improves quality of life in COPD patients who have chronic resting hypoxemia (room air Pao2 ≤ 55 mm Hg, or ≤ 59 mm Hg with signs of right-sided heart strain or polycythemia).
PULMONARY REHABILITATION
Pulmonary rehabilitation likely reduces hospital admissions by improving exercise capacity.13 A systematic review of six trials in 230 patients found that respiratory rehabilitation after an acute COPD exacerbation reduced the risk of COPD hospital admission (RR 0.26, 95% CI 0.12–0.54) and the risk of death (RR 0.45, 95% CI 0.22–0.91).13
OTHER INTERVENTIONS
Home noninvasive ventilator support reduced hospital and intensive care unit readmissions in select patients recurrently hospitalized for acidotic exacerbations of COPD in one small study.14
Long-term antibiotic therapy. Although there is evidence that azithromycin, taken daily for 1 year, decreases the frequency of COPD exacerbations,15 concern persists that this approach promotes antibiotic resistance, and the GOLD guidelines do not recommend routinely using antibiotics in patients with clinically stable COPD.6
Roflumilast. According to the GOLD guidelines, the phosphodiesterase-4 inhibitor roflumilast (Daliresp) may be useful in reducing exacerbations in patients who have an FEV1 less than 50% of predicted, chronic bronchitis, and frequent exacerbations.6
Referral. Patients who have severe recurrent COPD exacerbations despite appropriate therapy will likely benefit from referral to a pulmonary specialist for other options such as theophylline, lung-reduction surgery, and lung transplantation.
PATIENT EDUCATION AND OUTPATIENT COPD PROGRAMS
There is growing evidence that outpatient programs that provide education and medical support significantly reduce the rate of hospitalizations for COPD.16–18 Patient education includes symptom monitoring, early recognition of an exacerbation, appropriate use of inhalers and nebulizers, and advice on smoking cessation.16
On the other hand, a Veterans Administration randomized controlled trial was stopped early because of a higher rate of death in the group that underwent a comprehensive care-management program of COPD education, an action plan for identification and treatment of exacerbations, and scheduled proactive telephone calls for case management.19
Further study is needed to investigate the cost-effectiveness and safety of COPD management programs and whether to adopt such programs on a systematic level.
In conclusion, COPD patients require a comprehensive approach based on studied interventions. This may be achieved through systematic methods that allow each patient to benefit from all possible interventions appropriate for him or her. Hospitalization of COPD patients provides an excellent opportunity to implement this comprehensive approach.
We think so. Some strategies to reduce readmission rates, such as coordinating care and managing comorbidities, apply to chronic diseases in general, while others are disease-specific. To reduce the need for hospital readmission for chronic obstructive pulmonary disease (COPD), coordinated efforts involving both inpatient and outpatient care are necessary. This can be achieved by using a checklist before discharge (Table 1) and by implementing outpatient COPD programs that continue patient education and provide rapid access to medical support if needed.
There is room for improvement. COPD is common and expensive, with high rates of hospital readmission,1 and up to 70% of the money we spend on it goes for hospital care.2 No wonder then that the Centers for Medicare and Medicaid Services has now expanded its Readmissions Reduction Program to include acute COPD exacerbations.3 Yet in a retrospective study, Yip et al4 found that fewer than half of patients hospitalized with acute exacerbation of COPD received appropriate vaccinations, counseling on smoking cessation, and long-acting inhalers—all of which are on our checklist.4
The following interventions have been demonstrated to be useful in reducing COPD hospital admissions and the risk of death.
SMOKING CESSATION
Cigarette smoking is the most common and easily identifiable risk factor for COPD exacerbation.5
Au et al5 found that quitting smoking reduces the risk of COPD exacerbation (adjusted hazard ratio 0.78, 95% confidence interval [CI] 0.75–0.87), and the risk keeps decreasing the longer the patient stays off tobacco.5
Whether counseling hospitalized patients on smoking cessation reduces the COPD readmission rate has not been well studied. However, a meta-analysis of nine randomized controlled trials, two of which were done in the hospital, revealed higher abstinence rates in COPD patients who received extensive counseling on smoking cessation.7 For these reasons, hospitalized COPD patients who smoke should be strongly encouraged to quit.6
PNEUMOCOCCAL AND INFLUENZA VACCINATIONS
In a large retrospective study,8 pneumococcal vaccination was associated with a significantly lower risk of hospitalization for pneumonia in patients with chronic lung disease, including those with COPD (relative risk [RR] 0.57, 95% CI 0.38–0.84). The benefit was even greater with pneumococcal and influenza vaccinations during the influenza season (RR 0.28, 95% CI 0.14–0.58).
Randomized controlled trials indicate that influenza vaccination may reduce the rate of COPD exacerbations, especially in epidemic years when the proportion of exacerbations due to influenza is higher.9
Wongsurakiat et al10 found a significant reduction in the incidence of influenza-related acute respiratory illness in COPD patients in a well-designed randomized, placebo-controlled trial (RR 0.24, P = .005).10
Similarly, in another randomized controlled trial, pneumococcal vaccination was effective in preventing community-acquired pneumonia in COPD patients under age 65 and in those with severe airflow obstruction, although no statistically significant differences were found among other groups of patients with COPD.11
Therefore, influenza and pneumococcal vaccinations are recommended by major COPD guidelines, such as GOLD (Global Strategy for the Diagnosis, Management, and Prevention of Chronic Obstructive Lung Disease).6
INHALERS
Inhaler therapy is recommended based on COPD severity according to GOLD classification, and appropriate inhaler therapy with proper inhaler technique reduces the number of COPD exacerbations and hospitalizations.6
Long-acting beta-agonists and anticholinergics reduce the risk of COPD exacerbation and hospitalization and so are preferred over short-acting formulations except for patients in GOLD group A, ie, those who have few symptoms and are at low risk of exacerbations.6
Long-term treatment with inhaled corticosteroids with long-acting bronchodilators is recommended for patients at high risk of exacerbations (ie, those with two or more exacerbations in the previous year or a forced expiratory volume in 1 second [FEV1] less than 50% of predicted).6
OXYGEN THERAPY
Two older randomized controlled trials, the Nocturnal Oxygen Therapy Trial and the Medical Research Council study, reviewed by Stoller et al,12 provided clear evidence that oxygen therapy reduces the death rate and improves quality of life in COPD patients who have chronic resting hypoxemia (room air Pao2 ≤ 55 mm Hg, or ≤ 59 mm Hg with signs of right-sided heart strain or polycythemia).
PULMONARY REHABILITATION
Pulmonary rehabilitation likely reduces hospital admissions by improving exercise capacity.13 A systematic review of six trials in 230 patients found that respiratory rehabilitation after an acute COPD exacerbation reduced the risk of COPD hospital admission (RR 0.26, 95% CI 0.12–0.54) and the risk of death (RR 0.45, 95% CI 0.22–0.91).13
OTHER INTERVENTIONS
Home noninvasive ventilator support reduced hospital and intensive care unit readmissions in select patients recurrently hospitalized for acidotic exacerbations of COPD in one small study.14
Long-term antibiotic therapy. Although there is evidence that azithromycin, taken daily for 1 year, decreases the frequency of COPD exacerbations,15 concern persists that this approach promotes antibiotic resistance, and the GOLD guidelines do not recommend routinely using antibiotics in patients with clinically stable COPD.6
Roflumilast. According to the GOLD guidelines, the phosphodiesterase-4 inhibitor roflumilast (Daliresp) may be useful in reducing exacerbations in patients who have an FEV1 less than 50% of predicted, chronic bronchitis, and frequent exacerbations.6
Referral. Patients who have severe recurrent COPD exacerbations despite appropriate therapy will likely benefit from referral to a pulmonary specialist for other options such as theophylline, lung-reduction surgery, and lung transplantation.
PATIENT EDUCATION AND OUTPATIENT COPD PROGRAMS
There is growing evidence that outpatient programs that provide education and medical support significantly reduce the rate of hospitalizations for COPD.16–18 Patient education includes symptom monitoring, early recognition of an exacerbation, appropriate use of inhalers and nebulizers, and advice on smoking cessation.16
On the other hand, a Veterans Administration randomized controlled trial was stopped early because of a higher rate of death in the group that underwent a comprehensive care-management program of COPD education, an action plan for identification and treatment of exacerbations, and scheduled proactive telephone calls for case management.19
Further study is needed to investigate the cost-effectiveness and safety of COPD management programs and whether to adopt such programs on a systematic level.
In conclusion, COPD patients require a comprehensive approach based on studied interventions. This may be achieved through systematic methods that allow each patient to benefit from all possible interventions appropriate for him or her. Hospitalization of COPD patients provides an excellent opportunity to implement this comprehensive approach.
- Westert GP, Lagoe RJ, Keskimäki I, Leyland A, Murphy M. An international study of hospital readmissions and related utilization in Europe and the USA. Health Policy 2002; 61:269–278.
- Halpern MT, Stanford RH, Borker R. The burden of COPD in the USA: results from the Confronting COPD survey. Respir Med 2003; 97(suppl C):S81–S89.
- Centers for Medicare and Medicaid Services. Readmissions reduction program. www.cms.gov/Medicare/Medicare-Fee-for-Service-Payment/AcuteInpatientPPS/Readmissions-Reduction-Program.html. Accessed August 9, 2014.
- Yip NH, Yuen G, Lazar EJ, et al. Analysis of hospitalizations for COPD exacerbation: opportunities for improving care. COPD 2010; 7:85–92.
- Au DH, Bryson CL, Chien JW, et al. The effects of smoking cessation on the risk of chronic obstructive pulmonary disease exacerbations. J Gen Intern Med 2009; 24:457–463.
- Vestbo J, Hurd SS, Agustí AG, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med 2013; 187:347–365.
- Thabane MCOPD Working Group. Smoking cessation for patients with chronic obstructive pulmonary disease (COPD): an evidence-based analysis. Ont Health Technol Assess Ser 2012; 12:1–50.
- Nichol KL, Baken L, Wuorenma J, Nelson A. The health and economic benefits associated with pneumococcal vaccination of elderly persons with chronic lung disease. Arch Intern Med 1999; 159:2437–2442.
- Poole PJ, Chacko E, Wood-Baker RW, Cates CJ. Influenza vaccine for patients with chronic obstructive pulmonary disease. Cochrane Database Syst Rev 2006; 1:CD002733.
- Wongsurakiat P, Maranetra KN, Wasi C, Kositanont U, Dejsomritrutai W, Charoenratanakul S. Acute respiratory illness in patients with COPD and the effectiveness of influenza vaccination: a randomized controlled study. Chest 2004; 125:2011–2020.
- Alfageme I, Vazquez R, Reyes N, et al. Clinical efficacy of anti-pneumococcal vaccination in patients with COPD. Thorax 2006; 61:189–195.
- Stoller JK, Panos RJ, Krachman S, Doherty DE, Make B; Long-term Oxygen Treatment Trial Research Group. Oxygen therapy for patients with COPD: current evidence and the long-term oxygen treatment trial. Chest 2010; 138:179–187.
- Puhan MA, Scharplatz M, Troosters T, Steurer J. Respiratory rehabilitation after acute exacerbation of COPD may reduce risk for readmission and mortality—a systematic review. Respir Res 2005; 6:54.
- Tuggey JM, Plant PK, Elliott MW. Domiciliary non-invasive ventilation for recurrent acidotic exacerbations of COPD: an economic analysis. Thorax 2003; 58:867–871.
- Albert RK, Connett J, Bailey WC, et al; COPD Clinical Research Network. Azithromycin for prevention of exacerbations of COPD. N Engl J Med 2011; 365:689–698.
- Lawlor M, Kealy S, Agnew M, et al. Early discharge care with ongoing follow-up support may reduce hospital readmissions in COPD. Int J Chron Obstruct Pulmon Dis 2009; 4:55–60.
- Gadoury MA, Schwartzman K, Rouleau M, et al; Chronic Obstructive Pulmonary Disease axis of the Respiratory Health Network, Fonds de la Recherche en Santé du Québec (FRSQ). Self-management reduces both short- and long-term hospitalisation in COPD. Eur Respir J 2005; 26:853–857.
- Rice KL, Dewan N, Bloomfield HE, et al. Disease management program for chronic obstructive pulmonary disease: a randomized controlled trial. Am J Respir Crit Care Med 2010; 182:890–896.
- Fan VS, Gaziano JM, Lew R, et al. A comprehensive care management program to prevent chronic obstructive pulmonary disease hospitalizations: a randomized, controlled trial. Ann Intern Med 2012; 156:673–683.
- COPD Working Group. Noninvasive positive pressure ventilation for chronic respiratory failure patients with stable chronic obstructive pulmonary disease (COPD): an evidence-based analysis. Ont Health Technol Assess Ser 2012; 12( 9):1–51.
- Westert GP, Lagoe RJ, Keskimäki I, Leyland A, Murphy M. An international study of hospital readmissions and related utilization in Europe and the USA. Health Policy 2002; 61:269–278.
- Halpern MT, Stanford RH, Borker R. The burden of COPD in the USA: results from the Confronting COPD survey. Respir Med 2003; 97(suppl C):S81–S89.
- Centers for Medicare and Medicaid Services. Readmissions reduction program. www.cms.gov/Medicare/Medicare-Fee-for-Service-Payment/AcuteInpatientPPS/Readmissions-Reduction-Program.html. Accessed August 9, 2014.
- Yip NH, Yuen G, Lazar EJ, et al. Analysis of hospitalizations for COPD exacerbation: opportunities for improving care. COPD 2010; 7:85–92.
- Au DH, Bryson CL, Chien JW, et al. The effects of smoking cessation on the risk of chronic obstructive pulmonary disease exacerbations. J Gen Intern Med 2009; 24:457–463.
- Vestbo J, Hurd SS, Agustí AG, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med 2013; 187:347–365.
- Thabane MCOPD Working Group. Smoking cessation for patients with chronic obstructive pulmonary disease (COPD): an evidence-based analysis. Ont Health Technol Assess Ser 2012; 12:1–50.
- Nichol KL, Baken L, Wuorenma J, Nelson A. The health and economic benefits associated with pneumococcal vaccination of elderly persons with chronic lung disease. Arch Intern Med 1999; 159:2437–2442.
- Poole PJ, Chacko E, Wood-Baker RW, Cates CJ. Influenza vaccine for patients with chronic obstructive pulmonary disease. Cochrane Database Syst Rev 2006; 1:CD002733.
- Wongsurakiat P, Maranetra KN, Wasi C, Kositanont U, Dejsomritrutai W, Charoenratanakul S. Acute respiratory illness in patients with COPD and the effectiveness of influenza vaccination: a randomized controlled study. Chest 2004; 125:2011–2020.
- Alfageme I, Vazquez R, Reyes N, et al. Clinical efficacy of anti-pneumococcal vaccination in patients with COPD. Thorax 2006; 61:189–195.
- Stoller JK, Panos RJ, Krachman S, Doherty DE, Make B; Long-term Oxygen Treatment Trial Research Group. Oxygen therapy for patients with COPD: current evidence and the long-term oxygen treatment trial. Chest 2010; 138:179–187.
- Puhan MA, Scharplatz M, Troosters T, Steurer J. Respiratory rehabilitation after acute exacerbation of COPD may reduce risk for readmission and mortality—a systematic review. Respir Res 2005; 6:54.
- Tuggey JM, Plant PK, Elliott MW. Domiciliary non-invasive ventilation for recurrent acidotic exacerbations of COPD: an economic analysis. Thorax 2003; 58:867–871.
- Albert RK, Connett J, Bailey WC, et al; COPD Clinical Research Network. Azithromycin for prevention of exacerbations of COPD. N Engl J Med 2011; 365:689–698.
- Lawlor M, Kealy S, Agnew M, et al. Early discharge care with ongoing follow-up support may reduce hospital readmissions in COPD. Int J Chron Obstruct Pulmon Dis 2009; 4:55–60.
- Gadoury MA, Schwartzman K, Rouleau M, et al; Chronic Obstructive Pulmonary Disease axis of the Respiratory Health Network, Fonds de la Recherche en Santé du Québec (FRSQ). Self-management reduces both short- and long-term hospitalisation in COPD. Eur Respir J 2005; 26:853–857.
- Rice KL, Dewan N, Bloomfield HE, et al. Disease management program for chronic obstructive pulmonary disease: a randomized controlled trial. Am J Respir Crit Care Med 2010; 182:890–896.
- Fan VS, Gaziano JM, Lew R, et al. A comprehensive care management program to prevent chronic obstructive pulmonary disease hospitalizations: a randomized, controlled trial. Ann Intern Med 2012; 156:673–683.
- COPD Working Group. Noninvasive positive pressure ventilation for chronic respiratory failure patients with stable chronic obstructive pulmonary disease (COPD): an evidence-based analysis. Ont Health Technol Assess Ser 2012; 12( 9):1–51.
To improve our patients’ health, look beyond reducing readmissions
In this issue of the Cleveland Clinic Journal of Medicine, Drs. Ayache, Boyaji, and Pile share evidence-based strategies for reducing the risk of readmission for patients with acute exacerbations of chronic obstructive pulmonary disease (COPD).1 They emphasize standardizing practice by combining effective clinical management with appropriate patient education, communication, and postdischarge follow-up.
Reducing the rate of preventable hospital readmissions (as well as avoiding admissions in the first place) is the right thing to do for the patient. Moreover, broader adoption of the strategies that they outline in their article will be critical to the success of health care organizations in improving patient outcomes and navigating a rapidly evolving landscape of reimbursement and reporting changes associated with the Centers for Medicare and Medicaid Services (CMS) Readmissions Reduction Program. Hospital readmission rates, while imperfect measures of the quality of care, demonstrate opportunities to optimize transitions of care. Success in our efforts to improve the health of our patients will likely be aligned with reductions in preventable admissions and improved attention to care coordination.
HOSPITALS ARE PENALIZED FOR EXCESSIVE READMISSION RATES
With nearly 20% of Medicare beneficiaries being rehospitalized within 30 days of discharge, at a cost of $17 billion annually,2 Congress enacted the Hospital Readmissions Reduction Program3 as part of the Affordable Care Act (ACA) in 2012. The Centers for Medicare and Medicaid Services (CMS) had already been reporting the readmission rates for heart failure, acute myocardial infarction, and pneumonia since 2009 (www.medicare.gov/hospitalcompare). Building on this work, the Affordable Care Act implemented financial penalties against hospitals that had excessive rates of readmissions for these conditions.
The Affordable Care Act put 1% of a hospital’s Medicare base payment at risk for all inpatient diagnoses in 2013—not just the three listed here. The risk is 2% in 2014 and will rise to 3% in 2015. In its first year, more than 2,200 United States hospitals were penalized a total of approximately $280 million because of readmission rates above the national mean. Nearly 10% of hospitals incurred the maximum 1% penalty, and about 30% paid no penalty.
The Secretary of the Department of Health and Human Services has the authority to extend the Readmissions Reduction Program to additional high-volume or high-expenditure conditions, and the department has announced it will expand the program in October 2014 (fiscal year 2015) to include two additional conditions: elective hip or knee replacement and COPD.4 In both cases, CMS began by publicly reporting these rates before including them in the program. Additional readmission measures, including those for stroke and hospital-wide all-cause readmissions, are also publicly reported and receive increased attention but are not yet included in the Readmissions Reduction Program.
UNFAIRLY PENALIZING THOSE THAT SERVE THE POOR
Avoidable causes of readmissions include hospital-acquired infections and complications, inadequate medication reconciliation and management, poor communication and coordination of care among the members of the health care team, and suboptimal care transitions.5 But other important drivers of readmissions are outside of a hospital’s direct control. These include mental illness, lack of social support, and poverty.6
A criticism of the Readmissions Reduction Program is that it disproportionately penalizes hospitals that serve the poorest patients.7 Currently, CMS readmission risk models do not adjust for socioeconomic factors. Further, CMS responds to these concerns by noting that it does not want different outcome standards for poor patients, and that adjusting for these factors may conceal potential health care disparities in disadvantaged populations.
NEW MISSION FOR HOSPITALS: MITIGATE SOCIOECONOMIC BARRIERS
Effective programs to reduce hospital readmissions must address the clinical interventions and patient education needs in the COPD discharge checklist discussed by Ayache et al, but must also attempt to mitigate social disadvantages that drive up readmissions for patients at highest risk.
Are hospitals in a position to do this? Too often, it is assumed that patients have access to medications, transportation to follow-up appointments, and social support. Early identification of patients at highest risk of being affected by lack of these factors and innovative solutions for mitigating these risks are important considerations in our efforts to reduce hospital readmissions.
HOW MANY READMISSIONS ARE TRULY PREVENTABLE?
Experts disagree on how many readmissions are truly preventable. Readmission rates for the sickest patients treated at tertiary or academic medical centers may reflect high-quality care in well-managed patients who otherwise would have died during the index admission.8
In early studies, the Medicare Payment Advisory Commission estimated that up to three-quarters of readmissions are preventable.9 In contrast, studies that used clinical instead of administrative data suggest preventable readmissions make up as little as 12% of total readmissions.10
Regardless of the actual percentage, Medicare’s risk-adjustment model relies exclusively on administrative data that do not fully account for nonpreventable factors and do not completely address unrelated or planned rehospitalizations. CMS is attempting to address these issues with an expanded readmission algorithm that excludes more planned and unrelated readmissions from the penalty calculation.
Ironically, the current structure of the Readmissions Reduction Program does little to address its intended goal of eliminating the perverse financial incentives for hospitals and physicians to readmit patients. Payments are still episode-based and reward readmissions. The $280 million that CMS expects to receive from the program this year covers less than 5% of the nearly $12 billion attributed to preventable rehospitalizations.11
WHAT PATIENTS NEED, NOT WHAT SUITS PROVIDERS
Hospital readmission rates are publicly reported, but it is shortsighted to think about readmissions outside of the broader context of the “medical home.” One must consider the role of primary care providers before and after an index admission in addition to the role of postacute care providers for some patients after discharge. Neither is directly affected by the current penalty program, but both are critical to effective solutions and optimizing value-oriented care.
Readmission rates are suboptimal measures, as they address presumed failures of hospital transitions rather than measuring care coordination and providing meaningful incentives to coordinate care. Yes, there is much to do to ensure effective transitions from the hospital to home or postacute settings. But to truly transform health care and deliver value, shouldn’t we strive to redesign the work flow around what patients need rather than what suits providers?
This effort should focus on managing the conditions that bring patients to the hospital. Medical homes and optimizing chronic disease care can play pivotal roles in improving quality and reducing costs. Coordination of care and disease-management programs have led to cost reductions of 30% or more12 and have reduced admission rates by more than 10%.13 While the nation waits for health care reimbursement models to better reward patient quality outcomes and population health while reducing costs, we can use measures such as the Agency for Healthcare Research and Quality’s Prevention Quality Indicators to identify early interventions in the ambulatory care setting that can prevent admissions, complications, and exacerbation of disease.
Payers should also experiment with and promote innovative bundled-payment models such as Geisinger Health System’s ProvenCare program, which sets a fixed price for surgical procedures and up to 90 days of posthospital care, including readmission. These warranty-like programs overcome financial incentives to readmit patients in Medicare’s volume-based diagnosis-related group payment system.5
Re-engineering the delivery of care requires realigning resources to improve efficiency and effectiveness. In the short term, hospitals that successfully reduce readmission rates can expect reduced net reimbursements, as the penalties currently do not exceed the lost revenue of readmissions.
Reducing preventable readmissions is the right thing to do, but not all hospitalizations and rehospitalizations are avoidable. Many readmissions reflect appropriate and necessary care. The relentless focus on the readmission rate diverts attention and resources from more proactive solutions and innovative approaches for increasing health care safety, quality outcomes, and value.
Hospitals are caught between the volume and value paradigms. Payment programs that reward proactive disease management and care coordination will do the most to reduce health care costs and improve the quality of care. Hospitals have a responsibility to efficiently and effectively manage acute care and optimize handoffs to the next provider. Medicare’s payment policies do not do enough today to align the financial and quality-of-care incentives.
- Ayache MB, Boyaji S, Pile J. Can we reduce the risk of readmission for a patient with an exacerbation of COPD? Cleve Clin J Med 2014; 81:525–527.
- Jencks SF, Williams MV, Coleman EA. Rehospitalizations among patients in the Medicare fee-for-service program. N Engl J Med 2009; 360:1418–1428.
- Department of Health and Human Services. Medicare Program; Hospital Inpatient Prospective Payment Systems for Acute Care Hospitals and the Long-Term Care Hospital Prospective Payment System and FY 2012 Rates; Hospitals’ FTE Resident Caps for Graduate Medical Education Payment; Final Rule. Federal Register 2011; 76:51475–51846. www.gpo.gov/fdsys/pkg/FR-2011-08-18/pdf/2011-19719.pdf. Accessed August 5, 2014.
- Department of Health and Human Services. Medicare program; hospital inpatient prospective payment systems for acute care hospitals and the long term care; hospital prospective payment system and fiscal year 2014 rates; quality reporting requirements for specific providers; hospital conditions of participation; payment policies related to patient status; final rule. Federal Register 2013; 78:50495–51040. www.gpo.gov/fdsys/pkg/FR-2013-08-19/pdf/2013-18956.pdf. Accessed August 5, 2014.
- Berenson RA, Paulus RA, Kalman NS. Medicare’s readmissions-reduction program—a positive alternative. N Engl J Med 2012; 366:1364–1366.
- Joynt KE, Jha AK. Thirty-day readmissions—truth and consequences. N Engl J Med 2012; 366:1366–1369.
- Rau J. Medicare to penalize 2,217 hospitals for excess readmissions. Kaiser Health News 2012. www.kaiserhealthnews.org/stories/2012/august/13/medicare-hospitals-readmissions-penalties.aspx. Accessed August 5, 2014.
- Gorodeski EZ, Starling RC, Blackstone EH. Are all readmissions bad readmissions? N Engl J Med 2010; 363:297–298.
- Medicare Payment Advisory Commission. Report to the Congress: Promoting Greater Efficiency in Medicare, June 2007. www.medpac.gov/documents/jun07_entirereport.pdf. Accessed August 5, 2014.
- van Walraven C, Bennett C, Jennings A, Austin PC, Forster AJ. Proportion of hospital readmissions deemed avoidable: a systematic review. CMAJ 2011; 183:E391–E402.
- CMS Fee For Service IPPS Payment File, Fiscal Year 2014. cms.gov/Medicare/Medicare-Fee-for-Service-Payment/AcuteInpatientPPS/Downloads/FY_14_FR_Impact_File.zip. Accessed August 5, 2014.
- Dartmouth Medical School Center for the Evaluative Clinical Sciences. The Dartmouth Atlas of Health Care, 2006. www.dartmouthat-las.org/downloads/atlases/2006_Chronic_Care_Atlas.pdf. Accessed August 5, 2014.
- Gold W, Kongstvedt P. How broadening DM’s focus helped shrink one plan’s costs. Managed Care 2003. www.managedcaremag.com/archives/0311/0311.minnesota.html. Accessed August 5, 2014.
In this issue of the Cleveland Clinic Journal of Medicine, Drs. Ayache, Boyaji, and Pile share evidence-based strategies for reducing the risk of readmission for patients with acute exacerbations of chronic obstructive pulmonary disease (COPD).1 They emphasize standardizing practice by combining effective clinical management with appropriate patient education, communication, and postdischarge follow-up.
Reducing the rate of preventable hospital readmissions (as well as avoiding admissions in the first place) is the right thing to do for the patient. Moreover, broader adoption of the strategies that they outline in their article will be critical to the success of health care organizations in improving patient outcomes and navigating a rapidly evolving landscape of reimbursement and reporting changes associated with the Centers for Medicare and Medicaid Services (CMS) Readmissions Reduction Program. Hospital readmission rates, while imperfect measures of the quality of care, demonstrate opportunities to optimize transitions of care. Success in our efforts to improve the health of our patients will likely be aligned with reductions in preventable admissions and improved attention to care coordination.
HOSPITALS ARE PENALIZED FOR EXCESSIVE READMISSION RATES
With nearly 20% of Medicare beneficiaries being rehospitalized within 30 days of discharge, at a cost of $17 billion annually,2 Congress enacted the Hospital Readmissions Reduction Program3 as part of the Affordable Care Act (ACA) in 2012. The Centers for Medicare and Medicaid Services (CMS) had already been reporting the readmission rates for heart failure, acute myocardial infarction, and pneumonia since 2009 (www.medicare.gov/hospitalcompare). Building on this work, the Affordable Care Act implemented financial penalties against hospitals that had excessive rates of readmissions for these conditions.
The Affordable Care Act put 1% of a hospital’s Medicare base payment at risk for all inpatient diagnoses in 2013—not just the three listed here. The risk is 2% in 2014 and will rise to 3% in 2015. In its first year, more than 2,200 United States hospitals were penalized a total of approximately $280 million because of readmission rates above the national mean. Nearly 10% of hospitals incurred the maximum 1% penalty, and about 30% paid no penalty.
The Secretary of the Department of Health and Human Services has the authority to extend the Readmissions Reduction Program to additional high-volume or high-expenditure conditions, and the department has announced it will expand the program in October 2014 (fiscal year 2015) to include two additional conditions: elective hip or knee replacement and COPD.4 In both cases, CMS began by publicly reporting these rates before including them in the program. Additional readmission measures, including those for stroke and hospital-wide all-cause readmissions, are also publicly reported and receive increased attention but are not yet included in the Readmissions Reduction Program.
UNFAIRLY PENALIZING THOSE THAT SERVE THE POOR
Avoidable causes of readmissions include hospital-acquired infections and complications, inadequate medication reconciliation and management, poor communication and coordination of care among the members of the health care team, and suboptimal care transitions.5 But other important drivers of readmissions are outside of a hospital’s direct control. These include mental illness, lack of social support, and poverty.6
A criticism of the Readmissions Reduction Program is that it disproportionately penalizes hospitals that serve the poorest patients.7 Currently, CMS readmission risk models do not adjust for socioeconomic factors. Further, CMS responds to these concerns by noting that it does not want different outcome standards for poor patients, and that adjusting for these factors may conceal potential health care disparities in disadvantaged populations.
NEW MISSION FOR HOSPITALS: MITIGATE SOCIOECONOMIC BARRIERS
Effective programs to reduce hospital readmissions must address the clinical interventions and patient education needs in the COPD discharge checklist discussed by Ayache et al, but must also attempt to mitigate social disadvantages that drive up readmissions for patients at highest risk.
Are hospitals in a position to do this? Too often, it is assumed that patients have access to medications, transportation to follow-up appointments, and social support. Early identification of patients at highest risk of being affected by lack of these factors and innovative solutions for mitigating these risks are important considerations in our efforts to reduce hospital readmissions.
HOW MANY READMISSIONS ARE TRULY PREVENTABLE?
Experts disagree on how many readmissions are truly preventable. Readmission rates for the sickest patients treated at tertiary or academic medical centers may reflect high-quality care in well-managed patients who otherwise would have died during the index admission.8
In early studies, the Medicare Payment Advisory Commission estimated that up to three-quarters of readmissions are preventable.9 In contrast, studies that used clinical instead of administrative data suggest preventable readmissions make up as little as 12% of total readmissions.10
Regardless of the actual percentage, Medicare’s risk-adjustment model relies exclusively on administrative data that do not fully account for nonpreventable factors and do not completely address unrelated or planned rehospitalizations. CMS is attempting to address these issues with an expanded readmission algorithm that excludes more planned and unrelated readmissions from the penalty calculation.
Ironically, the current structure of the Readmissions Reduction Program does little to address its intended goal of eliminating the perverse financial incentives for hospitals and physicians to readmit patients. Payments are still episode-based and reward readmissions. The $280 million that CMS expects to receive from the program this year covers less than 5% of the nearly $12 billion attributed to preventable rehospitalizations.11
WHAT PATIENTS NEED, NOT WHAT SUITS PROVIDERS
Hospital readmission rates are publicly reported, but it is shortsighted to think about readmissions outside of the broader context of the “medical home.” One must consider the role of primary care providers before and after an index admission in addition to the role of postacute care providers for some patients after discharge. Neither is directly affected by the current penalty program, but both are critical to effective solutions and optimizing value-oriented care.
Readmission rates are suboptimal measures, as they address presumed failures of hospital transitions rather than measuring care coordination and providing meaningful incentives to coordinate care. Yes, there is much to do to ensure effective transitions from the hospital to home or postacute settings. But to truly transform health care and deliver value, shouldn’t we strive to redesign the work flow around what patients need rather than what suits providers?
This effort should focus on managing the conditions that bring patients to the hospital. Medical homes and optimizing chronic disease care can play pivotal roles in improving quality and reducing costs. Coordination of care and disease-management programs have led to cost reductions of 30% or more12 and have reduced admission rates by more than 10%.13 While the nation waits for health care reimbursement models to better reward patient quality outcomes and population health while reducing costs, we can use measures such as the Agency for Healthcare Research and Quality’s Prevention Quality Indicators to identify early interventions in the ambulatory care setting that can prevent admissions, complications, and exacerbation of disease.
Payers should also experiment with and promote innovative bundled-payment models such as Geisinger Health System’s ProvenCare program, which sets a fixed price for surgical procedures and up to 90 days of posthospital care, including readmission. These warranty-like programs overcome financial incentives to readmit patients in Medicare’s volume-based diagnosis-related group payment system.5
Re-engineering the delivery of care requires realigning resources to improve efficiency and effectiveness. In the short term, hospitals that successfully reduce readmission rates can expect reduced net reimbursements, as the penalties currently do not exceed the lost revenue of readmissions.
Reducing preventable readmissions is the right thing to do, but not all hospitalizations and rehospitalizations are avoidable. Many readmissions reflect appropriate and necessary care. The relentless focus on the readmission rate diverts attention and resources from more proactive solutions and innovative approaches for increasing health care safety, quality outcomes, and value.
Hospitals are caught between the volume and value paradigms. Payment programs that reward proactive disease management and care coordination will do the most to reduce health care costs and improve the quality of care. Hospitals have a responsibility to efficiently and effectively manage acute care and optimize handoffs to the next provider. Medicare’s payment policies do not do enough today to align the financial and quality-of-care incentives.
In this issue of the Cleveland Clinic Journal of Medicine, Drs. Ayache, Boyaji, and Pile share evidence-based strategies for reducing the risk of readmission for patients with acute exacerbations of chronic obstructive pulmonary disease (COPD).1 They emphasize standardizing practice by combining effective clinical management with appropriate patient education, communication, and postdischarge follow-up.
Reducing the rate of preventable hospital readmissions (as well as avoiding admissions in the first place) is the right thing to do for the patient. Moreover, broader adoption of the strategies that they outline in their article will be critical to the success of health care organizations in improving patient outcomes and navigating a rapidly evolving landscape of reimbursement and reporting changes associated with the Centers for Medicare and Medicaid Services (CMS) Readmissions Reduction Program. Hospital readmission rates, while imperfect measures of the quality of care, demonstrate opportunities to optimize transitions of care. Success in our efforts to improve the health of our patients will likely be aligned with reductions in preventable admissions and improved attention to care coordination.
HOSPITALS ARE PENALIZED FOR EXCESSIVE READMISSION RATES
With nearly 20% of Medicare beneficiaries being rehospitalized within 30 days of discharge, at a cost of $17 billion annually,2 Congress enacted the Hospital Readmissions Reduction Program3 as part of the Affordable Care Act (ACA) in 2012. The Centers for Medicare and Medicaid Services (CMS) had already been reporting the readmission rates for heart failure, acute myocardial infarction, and pneumonia since 2009 (www.medicare.gov/hospitalcompare). Building on this work, the Affordable Care Act implemented financial penalties against hospitals that had excessive rates of readmissions for these conditions.
The Affordable Care Act put 1% of a hospital’s Medicare base payment at risk for all inpatient diagnoses in 2013—not just the three listed here. The risk is 2% in 2014 and will rise to 3% in 2015. In its first year, more than 2,200 United States hospitals were penalized a total of approximately $280 million because of readmission rates above the national mean. Nearly 10% of hospitals incurred the maximum 1% penalty, and about 30% paid no penalty.
The Secretary of the Department of Health and Human Services has the authority to extend the Readmissions Reduction Program to additional high-volume or high-expenditure conditions, and the department has announced it will expand the program in October 2014 (fiscal year 2015) to include two additional conditions: elective hip or knee replacement and COPD.4 In both cases, CMS began by publicly reporting these rates before including them in the program. Additional readmission measures, including those for stroke and hospital-wide all-cause readmissions, are also publicly reported and receive increased attention but are not yet included in the Readmissions Reduction Program.
UNFAIRLY PENALIZING THOSE THAT SERVE THE POOR
Avoidable causes of readmissions include hospital-acquired infections and complications, inadequate medication reconciliation and management, poor communication and coordination of care among the members of the health care team, and suboptimal care transitions.5 But other important drivers of readmissions are outside of a hospital’s direct control. These include mental illness, lack of social support, and poverty.6
A criticism of the Readmissions Reduction Program is that it disproportionately penalizes hospitals that serve the poorest patients.7 Currently, CMS readmission risk models do not adjust for socioeconomic factors. Further, CMS responds to these concerns by noting that it does not want different outcome standards for poor patients, and that adjusting for these factors may conceal potential health care disparities in disadvantaged populations.
NEW MISSION FOR HOSPITALS: MITIGATE SOCIOECONOMIC BARRIERS
Effective programs to reduce hospital readmissions must address the clinical interventions and patient education needs in the COPD discharge checklist discussed by Ayache et al, but must also attempt to mitigate social disadvantages that drive up readmissions for patients at highest risk.
Are hospitals in a position to do this? Too often, it is assumed that patients have access to medications, transportation to follow-up appointments, and social support. Early identification of patients at highest risk of being affected by lack of these factors and innovative solutions for mitigating these risks are important considerations in our efforts to reduce hospital readmissions.
HOW MANY READMISSIONS ARE TRULY PREVENTABLE?
Experts disagree on how many readmissions are truly preventable. Readmission rates for the sickest patients treated at tertiary or academic medical centers may reflect high-quality care in well-managed patients who otherwise would have died during the index admission.8
In early studies, the Medicare Payment Advisory Commission estimated that up to three-quarters of readmissions are preventable.9 In contrast, studies that used clinical instead of administrative data suggest preventable readmissions make up as little as 12% of total readmissions.10
Regardless of the actual percentage, Medicare’s risk-adjustment model relies exclusively on administrative data that do not fully account for nonpreventable factors and do not completely address unrelated or planned rehospitalizations. CMS is attempting to address these issues with an expanded readmission algorithm that excludes more planned and unrelated readmissions from the penalty calculation.
Ironically, the current structure of the Readmissions Reduction Program does little to address its intended goal of eliminating the perverse financial incentives for hospitals and physicians to readmit patients. Payments are still episode-based and reward readmissions. The $280 million that CMS expects to receive from the program this year covers less than 5% of the nearly $12 billion attributed to preventable rehospitalizations.11
WHAT PATIENTS NEED, NOT WHAT SUITS PROVIDERS
Hospital readmission rates are publicly reported, but it is shortsighted to think about readmissions outside of the broader context of the “medical home.” One must consider the role of primary care providers before and after an index admission in addition to the role of postacute care providers for some patients after discharge. Neither is directly affected by the current penalty program, but both are critical to effective solutions and optimizing value-oriented care.
Readmission rates are suboptimal measures, as they address presumed failures of hospital transitions rather than measuring care coordination and providing meaningful incentives to coordinate care. Yes, there is much to do to ensure effective transitions from the hospital to home or postacute settings. But to truly transform health care and deliver value, shouldn’t we strive to redesign the work flow around what patients need rather than what suits providers?
This effort should focus on managing the conditions that bring patients to the hospital. Medical homes and optimizing chronic disease care can play pivotal roles in improving quality and reducing costs. Coordination of care and disease-management programs have led to cost reductions of 30% or more12 and have reduced admission rates by more than 10%.13 While the nation waits for health care reimbursement models to better reward patient quality outcomes and population health while reducing costs, we can use measures such as the Agency for Healthcare Research and Quality’s Prevention Quality Indicators to identify early interventions in the ambulatory care setting that can prevent admissions, complications, and exacerbation of disease.
Payers should also experiment with and promote innovative bundled-payment models such as Geisinger Health System’s ProvenCare program, which sets a fixed price for surgical procedures and up to 90 days of posthospital care, including readmission. These warranty-like programs overcome financial incentives to readmit patients in Medicare’s volume-based diagnosis-related group payment system.5
Re-engineering the delivery of care requires realigning resources to improve efficiency and effectiveness. In the short term, hospitals that successfully reduce readmission rates can expect reduced net reimbursements, as the penalties currently do not exceed the lost revenue of readmissions.
Reducing preventable readmissions is the right thing to do, but not all hospitalizations and rehospitalizations are avoidable. Many readmissions reflect appropriate and necessary care. The relentless focus on the readmission rate diverts attention and resources from more proactive solutions and innovative approaches for increasing health care safety, quality outcomes, and value.
Hospitals are caught between the volume and value paradigms. Payment programs that reward proactive disease management and care coordination will do the most to reduce health care costs and improve the quality of care. Hospitals have a responsibility to efficiently and effectively manage acute care and optimize handoffs to the next provider. Medicare’s payment policies do not do enough today to align the financial and quality-of-care incentives.
- Ayache MB, Boyaji S, Pile J. Can we reduce the risk of readmission for a patient with an exacerbation of COPD? Cleve Clin J Med 2014; 81:525–527.
- Jencks SF, Williams MV, Coleman EA. Rehospitalizations among patients in the Medicare fee-for-service program. N Engl J Med 2009; 360:1418–1428.
- Department of Health and Human Services. Medicare Program; Hospital Inpatient Prospective Payment Systems for Acute Care Hospitals and the Long-Term Care Hospital Prospective Payment System and FY 2012 Rates; Hospitals’ FTE Resident Caps for Graduate Medical Education Payment; Final Rule. Federal Register 2011; 76:51475–51846. www.gpo.gov/fdsys/pkg/FR-2011-08-18/pdf/2011-19719.pdf. Accessed August 5, 2014.
- Department of Health and Human Services. Medicare program; hospital inpatient prospective payment systems for acute care hospitals and the long term care; hospital prospective payment system and fiscal year 2014 rates; quality reporting requirements for specific providers; hospital conditions of participation; payment policies related to patient status; final rule. Federal Register 2013; 78:50495–51040. www.gpo.gov/fdsys/pkg/FR-2013-08-19/pdf/2013-18956.pdf. Accessed August 5, 2014.
- Berenson RA, Paulus RA, Kalman NS. Medicare’s readmissions-reduction program—a positive alternative. N Engl J Med 2012; 366:1364–1366.
- Joynt KE, Jha AK. Thirty-day readmissions—truth and consequences. N Engl J Med 2012; 366:1366–1369.
- Rau J. Medicare to penalize 2,217 hospitals for excess readmissions. Kaiser Health News 2012. www.kaiserhealthnews.org/stories/2012/august/13/medicare-hospitals-readmissions-penalties.aspx. Accessed August 5, 2014.
- Gorodeski EZ, Starling RC, Blackstone EH. Are all readmissions bad readmissions? N Engl J Med 2010; 363:297–298.
- Medicare Payment Advisory Commission. Report to the Congress: Promoting Greater Efficiency in Medicare, June 2007. www.medpac.gov/documents/jun07_entirereport.pdf. Accessed August 5, 2014.
- van Walraven C, Bennett C, Jennings A, Austin PC, Forster AJ. Proportion of hospital readmissions deemed avoidable: a systematic review. CMAJ 2011; 183:E391–E402.
- CMS Fee For Service IPPS Payment File, Fiscal Year 2014. cms.gov/Medicare/Medicare-Fee-for-Service-Payment/AcuteInpatientPPS/Downloads/FY_14_FR_Impact_File.zip. Accessed August 5, 2014.
- Dartmouth Medical School Center for the Evaluative Clinical Sciences. The Dartmouth Atlas of Health Care, 2006. www.dartmouthat-las.org/downloads/atlases/2006_Chronic_Care_Atlas.pdf. Accessed August 5, 2014.
- Gold W, Kongstvedt P. How broadening DM’s focus helped shrink one plan’s costs. Managed Care 2003. www.managedcaremag.com/archives/0311/0311.minnesota.html. Accessed August 5, 2014.
- Ayache MB, Boyaji S, Pile J. Can we reduce the risk of readmission for a patient with an exacerbation of COPD? Cleve Clin J Med 2014; 81:525–527.
- Jencks SF, Williams MV, Coleman EA. Rehospitalizations among patients in the Medicare fee-for-service program. N Engl J Med 2009; 360:1418–1428.
- Department of Health and Human Services. Medicare Program; Hospital Inpatient Prospective Payment Systems for Acute Care Hospitals and the Long-Term Care Hospital Prospective Payment System and FY 2012 Rates; Hospitals’ FTE Resident Caps for Graduate Medical Education Payment; Final Rule. Federal Register 2011; 76:51475–51846. www.gpo.gov/fdsys/pkg/FR-2011-08-18/pdf/2011-19719.pdf. Accessed August 5, 2014.
- Department of Health and Human Services. Medicare program; hospital inpatient prospective payment systems for acute care hospitals and the long term care; hospital prospective payment system and fiscal year 2014 rates; quality reporting requirements for specific providers; hospital conditions of participation; payment policies related to patient status; final rule. Federal Register 2013; 78:50495–51040. www.gpo.gov/fdsys/pkg/FR-2013-08-19/pdf/2013-18956.pdf. Accessed August 5, 2014.
- Berenson RA, Paulus RA, Kalman NS. Medicare’s readmissions-reduction program—a positive alternative. N Engl J Med 2012; 366:1364–1366.
- Joynt KE, Jha AK. Thirty-day readmissions—truth and consequences. N Engl J Med 2012; 366:1366–1369.
- Rau J. Medicare to penalize 2,217 hospitals for excess readmissions. Kaiser Health News 2012. www.kaiserhealthnews.org/stories/2012/august/13/medicare-hospitals-readmissions-penalties.aspx. Accessed August 5, 2014.
- Gorodeski EZ, Starling RC, Blackstone EH. Are all readmissions bad readmissions? N Engl J Med 2010; 363:297–298.
- Medicare Payment Advisory Commission. Report to the Congress: Promoting Greater Efficiency in Medicare, June 2007. www.medpac.gov/documents/jun07_entirereport.pdf. Accessed August 5, 2014.
- van Walraven C, Bennett C, Jennings A, Austin PC, Forster AJ. Proportion of hospital readmissions deemed avoidable: a systematic review. CMAJ 2011; 183:E391–E402.
- CMS Fee For Service IPPS Payment File, Fiscal Year 2014. cms.gov/Medicare/Medicare-Fee-for-Service-Payment/AcuteInpatientPPS/Downloads/FY_14_FR_Impact_File.zip. Accessed August 5, 2014.
- Dartmouth Medical School Center for the Evaluative Clinical Sciences. The Dartmouth Atlas of Health Care, 2006. www.dartmouthat-las.org/downloads/atlases/2006_Chronic_Care_Atlas.pdf. Accessed August 5, 2014.
- Gold W, Kongstvedt P. How broadening DM’s focus helped shrink one plan’s costs. Managed Care 2003. www.managedcaremag.com/archives/0311/0311.minnesota.html. Accessed August 5, 2014.
Erythema and atrophy on the tongue
A 26-year-old woman was referred to the dermatology department with a 6-month history of a painful burning sensation on the tongue. Examination revealed a reddish, atrophic area on the dorsum of the tongue (Figure 1).
She had been treated unsuccessfully with topical antifungal drugs (clotrimazole and nystatin) for a presumed diagnosis of oral candidiasis. Otherwise, her medical history was notable only for occasional episodes of epigastric pain. She did not smoke or drink alcohol.
Fungal culture and oral exfoliative cytology studies were negative.
Laboratory results:
- Red blood cell count 3.9 × 1012/L (reference range 4.2–5.4)
- Hemoglobin 11.3 g/dL (12–16)
- Mean corpuscular volume 92 fL (80–99)
- Mean corpuscular hemoglobin 29 pg (27–34)
- Iron 14 μg/dL (37–145),
- Vitamin B12 119 pg/dL (250–900)
- Zinc 33 μg/dL (66–110)
- Serum gastric parietal cell antibody positive
- Serum creatinine and liver enzyme tests were normal.
Biopsy of the gastric mucosa revealed severe atrophic gastritis, so the possibility of atrophy related to gastroesophageal reflux was considered. But the laboratory results and the patient’s presentation pointed to iron deficiency and pernicious anemia (due to deficiency of vitamin B12). Zinc deficiency is associated with oral burning but not atrophic glossitis.
Based on the patient’s symptoms and the testing results, she was given the diagnosis of atrophic glossitis. She was treated with oral iron supplementation, intramuscular injections of vitamin B12, and oral zinc supplementation. The glossitis resolved, and the gastric symptoms improved within 2 months, thus supporting our diagnosis of atrophic glossitis.
ATROPHIC GLOSSITIS
The diagnosis of abnormalities of the tongue requires a thorough history, including onset and duration, antecedent symptoms, and tobacco and alcohol use. Examination of tongue morphology is also important.1 Tongue abnormalities related to tobacco use and to alcohol use include leukoplakia, erythroplakia, oral submucosal fibrosis, lichen planus, and oral squamous cell carcinoma.
Atrophic glossitis is often linked to an underlying nutritional deficiency of iron, folic acid, vitamin B12, riboflavin, or niacin, although other nutritional deficiencies can be implicated. As noted, zinc deficiency can cause oral burning but not atrophic glossitis, and it resolves with correction of the underlying deficiency.2 Cobalamin deficiency is the main cause of atrophic glossitis.
As our patient’s presentation illustrated, oral symptoms can be multifactorial. Oral conditions may be an early clinical manifestation of a nutritional deficiency, but they can also reflect an alteration of the gastric mucosa3; a bacterial, viral, or fungal infection; neoplastic disease; autoimmune disease; endocrine disorder; local mechanical trauma; exposure to an irritant; or an allergic reaction.2
- Reamy BV, Derby R, Bunt CW. Common tongue conditions in primary care. Am Fam Physician 2010; 81:627–634.
- Chi AC, Neville BW, Krayer JW, Gonsalves WC. Oral manifestations of systemic disease. Am Fam Physician 2010; 82:1381–1388.
- Sun A, Lin HP, Wang YP, Chiang CP. Significant association of deficiency of hemoglobin, iron and vitamin B12, high homocysteine level, and gastric parietal cell antibody positivity with atrophic glossitis. J Oral Pathol Med 2012; 41:500–504.
A 26-year-old woman was referred to the dermatology department with a 6-month history of a painful burning sensation on the tongue. Examination revealed a reddish, atrophic area on the dorsum of the tongue (Figure 1).
She had been treated unsuccessfully with topical antifungal drugs (clotrimazole and nystatin) for a presumed diagnosis of oral candidiasis. Otherwise, her medical history was notable only for occasional episodes of epigastric pain. She did not smoke or drink alcohol.
Fungal culture and oral exfoliative cytology studies were negative.
Laboratory results:
- Red blood cell count 3.9 × 1012/L (reference range 4.2–5.4)
- Hemoglobin 11.3 g/dL (12–16)
- Mean corpuscular volume 92 fL (80–99)
- Mean corpuscular hemoglobin 29 pg (27–34)
- Iron 14 μg/dL (37–145),
- Vitamin B12 119 pg/dL (250–900)
- Zinc 33 μg/dL (66–110)
- Serum gastric parietal cell antibody positive
- Serum creatinine and liver enzyme tests were normal.
Biopsy of the gastric mucosa revealed severe atrophic gastritis, so the possibility of atrophy related to gastroesophageal reflux was considered. But the laboratory results and the patient’s presentation pointed to iron deficiency and pernicious anemia (due to deficiency of vitamin B12). Zinc deficiency is associated with oral burning but not atrophic glossitis.
Based on the patient’s symptoms and the testing results, she was given the diagnosis of atrophic glossitis. She was treated with oral iron supplementation, intramuscular injections of vitamin B12, and oral zinc supplementation. The glossitis resolved, and the gastric symptoms improved within 2 months, thus supporting our diagnosis of atrophic glossitis.
ATROPHIC GLOSSITIS
The diagnosis of abnormalities of the tongue requires a thorough history, including onset and duration, antecedent symptoms, and tobacco and alcohol use. Examination of tongue morphology is also important.1 Tongue abnormalities related to tobacco use and to alcohol use include leukoplakia, erythroplakia, oral submucosal fibrosis, lichen planus, and oral squamous cell carcinoma.
Atrophic glossitis is often linked to an underlying nutritional deficiency of iron, folic acid, vitamin B12, riboflavin, or niacin, although other nutritional deficiencies can be implicated. As noted, zinc deficiency can cause oral burning but not atrophic glossitis, and it resolves with correction of the underlying deficiency.2 Cobalamin deficiency is the main cause of atrophic glossitis.
As our patient’s presentation illustrated, oral symptoms can be multifactorial. Oral conditions may be an early clinical manifestation of a nutritional deficiency, but they can also reflect an alteration of the gastric mucosa3; a bacterial, viral, or fungal infection; neoplastic disease; autoimmune disease; endocrine disorder; local mechanical trauma; exposure to an irritant; or an allergic reaction.2
A 26-year-old woman was referred to the dermatology department with a 6-month history of a painful burning sensation on the tongue. Examination revealed a reddish, atrophic area on the dorsum of the tongue (Figure 1).
She had been treated unsuccessfully with topical antifungal drugs (clotrimazole and nystatin) for a presumed diagnosis of oral candidiasis. Otherwise, her medical history was notable only for occasional episodes of epigastric pain. She did not smoke or drink alcohol.
Fungal culture and oral exfoliative cytology studies were negative.
Laboratory results:
- Red blood cell count 3.9 × 1012/L (reference range 4.2–5.4)
- Hemoglobin 11.3 g/dL (12–16)
- Mean corpuscular volume 92 fL (80–99)
- Mean corpuscular hemoglobin 29 pg (27–34)
- Iron 14 μg/dL (37–145),
- Vitamin B12 119 pg/dL (250–900)
- Zinc 33 μg/dL (66–110)
- Serum gastric parietal cell antibody positive
- Serum creatinine and liver enzyme tests were normal.
Biopsy of the gastric mucosa revealed severe atrophic gastritis, so the possibility of atrophy related to gastroesophageal reflux was considered. But the laboratory results and the patient’s presentation pointed to iron deficiency and pernicious anemia (due to deficiency of vitamin B12). Zinc deficiency is associated with oral burning but not atrophic glossitis.
Based on the patient’s symptoms and the testing results, she was given the diagnosis of atrophic glossitis. She was treated with oral iron supplementation, intramuscular injections of vitamin B12, and oral zinc supplementation. The glossitis resolved, and the gastric symptoms improved within 2 months, thus supporting our diagnosis of atrophic glossitis.
ATROPHIC GLOSSITIS
The diagnosis of abnormalities of the tongue requires a thorough history, including onset and duration, antecedent symptoms, and tobacco and alcohol use. Examination of tongue morphology is also important.1 Tongue abnormalities related to tobacco use and to alcohol use include leukoplakia, erythroplakia, oral submucosal fibrosis, lichen planus, and oral squamous cell carcinoma.
Atrophic glossitis is often linked to an underlying nutritional deficiency of iron, folic acid, vitamin B12, riboflavin, or niacin, although other nutritional deficiencies can be implicated. As noted, zinc deficiency can cause oral burning but not atrophic glossitis, and it resolves with correction of the underlying deficiency.2 Cobalamin deficiency is the main cause of atrophic glossitis.
As our patient’s presentation illustrated, oral symptoms can be multifactorial. Oral conditions may be an early clinical manifestation of a nutritional deficiency, but they can also reflect an alteration of the gastric mucosa3; a bacterial, viral, or fungal infection; neoplastic disease; autoimmune disease; endocrine disorder; local mechanical trauma; exposure to an irritant; or an allergic reaction.2
- Reamy BV, Derby R, Bunt CW. Common tongue conditions in primary care. Am Fam Physician 2010; 81:627–634.
- Chi AC, Neville BW, Krayer JW, Gonsalves WC. Oral manifestations of systemic disease. Am Fam Physician 2010; 82:1381–1388.
- Sun A, Lin HP, Wang YP, Chiang CP. Significant association of deficiency of hemoglobin, iron and vitamin B12, high homocysteine level, and gastric parietal cell antibody positivity with atrophic glossitis. J Oral Pathol Med 2012; 41:500–504.
- Reamy BV, Derby R, Bunt CW. Common tongue conditions in primary care. Am Fam Physician 2010; 81:627–634.
- Chi AC, Neville BW, Krayer JW, Gonsalves WC. Oral manifestations of systemic disease. Am Fam Physician 2010; 82:1381–1388.
- Sun A, Lin HP, Wang YP, Chiang CP. Significant association of deficiency of hemoglobin, iron and vitamin B12, high homocysteine level, and gastric parietal cell antibody positivity with atrophic glossitis. J Oral Pathol Med 2012; 41:500–504.
'Allergic to the sun'
A 54-year-old white man presents to the emergency department with burning pain in his left upper arm for the past 2 to 3 days. His medical history includes seizure disorder, for which he takes levetiracetam (Keppra); hypertension, for which he takes metoprolol succinate (Toprol); and in the remote past, a gunshot wound to the head that left his right arm with residual contracture and weakness.
He says he is homeless, has been “allergic to the sun for a while,” and has had dark-colored urine and intermittent abdominal pain. He states that he does not use illicit substances but that he drinks 6 to 12 beers per night and smokes 1 pack of cigarettes per day.
Initial vital signs:
- Temperature 37.7°C (99.9°F)
- Blood pressure 217/114 mm Hg
- Heart rate 82 bpm
- Respiratory rate 18 per minute
- Capillary oxygen saturation 98% while breathing room air.
On examination, his right arm is significantly weak and contracted. His left arm has decreased sensation to pinprick and light touch from elbow to fingers. His face and both arms show hyperpigmentation alternating with atrophic scarring, which also affects his lips. There is no overt mucosal involvement. His hands and forearms have a sclerotic texture and patchy hair loss. Several small bullae are present on the dorsum of the left forearm and hand. There is a 6-inch, irregular, open lesion on the left forearm and a 1-inch lesion on the left hand (Figure 1).
Initial laboratory studies show:
- Chemistries and complete blood cell count within normal limits
- Platelet count 305 × 109/L (reference range 150–350)
- Orange-colored urine
- Hepatitis C virus (HCV) antibody positive (new finding)
- Human immunodeficiency virus antibody, hepatitis B surface antigen, and antinuclear antibody negative
- Phenytoin and urine drug screen negative
- Aspartate aminotransferase 70 U/L (reference range 5–34)
- Alanine aminotransferase 73 U/L (reference range 0–55)
- Prothrombin time 10.8 seconds (reference range 8.3–13.0), international normalized ratio 0.98 (reference range 0.8–1.2)
- Iron studies within normal limits.
The patient is admitted to the hospital and is started on cefazolin and clindamycin. Urine is collected for a porphyrin screen, and punch-biopsy samples from the forearms are sent for study. Ultrasonography shows splenomegaly, as well as increased echogenicity of the liver without structural abnormalities. Blood and urine cultures, drawn upon admission, are negative by discharge.
Pathologic study of the punch-biopsy specimens (Figure 2) shows the formation of subepidermal vesicles with extensive reticular and dermal fibrosis.
DIAGNOSIS: PORPHYRIA CUTANEA TARDA
Because of the patient’s history, examination, and pathology results, he was preliminarily diagnosed with porphyria cutanea tarda (PCT).1,2 The diagnosis was confirmed after he was discharged when his urine uroporphyrin level was found to be 157.5 μmol/mol of creatinine (reference range < 4) and his urine heptacarboxylporphyrin level was 118.0 μmol/mol of creatinine (reference range < 2).
This patient’s clinical presentation is classic for sporadic (ie, type 1) PCT. Sporadic PCT is an acquired deficiency of uroporphyrinogen decarboxylase, an enzyme that catalyzes the fifth step in heme metabolism.3 The deficiency of this enzyme is exclusively hepatic and is strongly associated with chronic hepatitis C infection. Mutations of the hemochromatosis gene (HFE), human immunodeficiency virus infection, alcohol use, and smoking are also risk factors.4 The prevalence in the United States is about 1:25,000; nearly 80% of cases are sporadic (type 1), and 20% are familial (type 2).5
Manifestations of PCT include photosensitive dermatitis, facial hypertrichosis, and orange urine.3 The photosensitivity dermatitis heals slowly and leads to sclerosis and hyperpigmentation.
Repeated phlebotomy is the first-line treatment, and hydroxychloroquine (Plaquenil) is the second-line treatment.6 Patients with PCT and hepatitis C should be considered for antiviral therapy according to standard guidelines. Treatment of hepatitis C may reduce the symptoms of PCT, even without a sustained viral response. However, not enough evidence exists to make treatment recommendations for this group.7
Because we were uncertain that the patient would return for follow-up, we did not start phlebotomy or treatment for hepatitis C. However, we did prescribe hydroxychloroquine 100 mg three times a week and instructed him to cover his skin when outside and to use effective sunblock. An outpatient visit was scheduled prior to discharge. Unfortunately, the patient was lost to follow-up.
Acknowledgment: The authors would like to personally thank Dr. Karen DeSouza from the University of Tennessee, Graduate School of Medicine, Department of Pathology, for her clinical expertise and kind advice.
- The University of Iowa, Department of Pathology, Laboratory Services Handbook. Porphyrins & Porphobilinogen, Urine (24 hr or random). www.healthcare.uiowa.edu/path_handbook/handbook/test2893.html. Accessed August 8, 2014.
- Maynard B, Peters MS. Histologic and immunofluorescence study of cutaneous porphyrias. J Cutan Pathol 1992; 19:40–47.
- Thunell S, Harper P. Porphyrins, porphyrin metabolism, porphyrias. III. Diagnosis, care and monitoring in porphyria cutanea tarda—suggestions for a handling programme. Scand J Clin Lab Invest 2000; 60:561–579.
- Lambrecht RW, Thapar M, Bonkovsky HL. Genetic aspects of porphyria cutanea tarda. Semin Liver Dis 2007; 27:99–108.
- Kushner JP, Barbuto AJ, Lee GR. An inherited enzymatic defect in porphyria cutanea tarda: decreased uroporphyrinogen decarboxylase activity. J Clin Invest 1976; 58:1089–1097.
- Singal AK, Kormos-Hallberg C, Lee C, et al. Low-dose hydroxychloroquine is as effective as phlebotomy in treatment of patients with porphyria cutanea tarda. Clin Gastroenterol Hepatol 2012; 10:1402–1409.
- Ryan Caballes F, Sendi H, Bonkovsky HL. Hepatitis C, porphyria cutanea tarda and liver iron: an update. Liver Int 2012; 32:880–893.
A 54-year-old white man presents to the emergency department with burning pain in his left upper arm for the past 2 to 3 days. His medical history includes seizure disorder, for which he takes levetiracetam (Keppra); hypertension, for which he takes metoprolol succinate (Toprol); and in the remote past, a gunshot wound to the head that left his right arm with residual contracture and weakness.
He says he is homeless, has been “allergic to the sun for a while,” and has had dark-colored urine and intermittent abdominal pain. He states that he does not use illicit substances but that he drinks 6 to 12 beers per night and smokes 1 pack of cigarettes per day.
Initial vital signs:
- Temperature 37.7°C (99.9°F)
- Blood pressure 217/114 mm Hg
- Heart rate 82 bpm
- Respiratory rate 18 per minute
- Capillary oxygen saturation 98% while breathing room air.
On examination, his right arm is significantly weak and contracted. His left arm has decreased sensation to pinprick and light touch from elbow to fingers. His face and both arms show hyperpigmentation alternating with atrophic scarring, which also affects his lips. There is no overt mucosal involvement. His hands and forearms have a sclerotic texture and patchy hair loss. Several small bullae are present on the dorsum of the left forearm and hand. There is a 6-inch, irregular, open lesion on the left forearm and a 1-inch lesion on the left hand (Figure 1).

Initial laboratory studies show:
- Chemistries and complete blood cell count within normal limits
- Platelet count 305 × 109/L (reference range 150–350)
- Orange-colored urine
- Hepatitis C virus (HCV) antibody positive (new finding)
- Human immunodeficiency virus antibody, hepatitis B surface antigen, and antinuclear antibody negative
- Phenytoin and urine drug screen negative
- Aspartate aminotransferase 70 U/L (reference range 5–34)
- Alanine aminotransferase 73 U/L (reference range 0–55)
- Prothrombin time 10.8 seconds (reference range 8.3–13.0), international normalized ratio 0.98 (reference range 0.8–1.2)
- Iron studies within normal limits.
The patient is admitted to the hospital and is started on cefazolin and clindamycin. Urine is collected for a porphyrin screen, and punch-biopsy samples from the forearms are sent for study. Ultrasonography shows splenomegaly, as well as increased echogenicity of the liver without structural abnormalities. Blood and urine cultures, drawn upon admission, are negative by discharge.
Pathologic study of the punch-biopsy specimens (Figure 2) shows the formation of subepidermal vesicles with extensive reticular and dermal fibrosis.

DIAGNOSIS: PORPHYRIA CUTANEA TARDA
Because of the patient’s history, examination, and pathology results, he was preliminarily diagnosed with porphyria cutanea tarda (PCT).1,2 The diagnosis was confirmed after he was discharged when his urine uroporphyrin level was found to be 157.5 μmol/mol of creatinine (reference range < 4) and his urine heptacarboxylporphyrin level was 118.0 μmol/mol of creatinine (reference range < 2).
This patient’s clinical presentation is classic for sporadic (ie, type 1) PCT. Sporadic PCT is an acquired deficiency of uroporphyrinogen decarboxylase, an enzyme that catalyzes the fifth step in heme metabolism.3 The deficiency of this enzyme is exclusively hepatic and is strongly associated with chronic hepatitis C infection. Mutations of the hemochromatosis gene (HFE), human immunodeficiency virus infection, alcohol use, and smoking are also risk factors.4 The prevalence in the United States is about 1:25,000; nearly 80% of cases are sporadic (type 1), and 20% are familial (type 2).5
Manifestations of PCT include photosensitive dermatitis, facial hypertrichosis, and orange urine.3 The photosensitivity dermatitis heals slowly and leads to sclerosis and hyperpigmentation.
Repeated phlebotomy is the first-line treatment, and hydroxychloroquine (Plaquenil) is the second-line treatment.6 Patients with PCT and hepatitis C should be considered for antiviral therapy according to standard guidelines. Treatment of hepatitis C may reduce the symptoms of PCT, even without a sustained viral response. However, not enough evidence exists to make treatment recommendations for this group.7
Because we were uncertain that the patient would return for follow-up, we did not start phlebotomy or treatment for hepatitis C. However, we did prescribe hydroxychloroquine 100 mg three times a week and instructed him to cover his skin when outside and to use effective sunblock. An outpatient visit was scheduled prior to discharge. Unfortunately, the patient was lost to follow-up.
Acknowledgment: The authors would like to personally thank Dr. Karen DeSouza from the University of Tennessee, Graduate School of Medicine, Department of Pathology, for her clinical expertise and kind advice.
A 54-year-old white man presents to the emergency department with burning pain in his left upper arm for the past 2 to 3 days. His medical history includes seizure disorder, for which he takes levetiracetam (Keppra); hypertension, for which he takes metoprolol succinate (Toprol); and in the remote past, a gunshot wound to the head that left his right arm with residual contracture and weakness.
He says he is homeless, has been “allergic to the sun for a while,” and has had dark-colored urine and intermittent abdominal pain. He states that he does not use illicit substances but that he drinks 6 to 12 beers per night and smokes 1 pack of cigarettes per day.
Initial vital signs:
- Temperature 37.7°C (99.9°F)
- Blood pressure 217/114 mm Hg
- Heart rate 82 bpm
- Respiratory rate 18 per minute
- Capillary oxygen saturation 98% while breathing room air.
On examination, his right arm is significantly weak and contracted. His left arm has decreased sensation to pinprick and light touch from elbow to fingers. His face and both arms show hyperpigmentation alternating with atrophic scarring, which also affects his lips. There is no overt mucosal involvement. His hands and forearms have a sclerotic texture and patchy hair loss. Several small bullae are present on the dorsum of the left forearm and hand. There is a 6-inch, irregular, open lesion on the left forearm and a 1-inch lesion on the left hand (Figure 1).

Initial laboratory studies show:
- Chemistries and complete blood cell count within normal limits
- Platelet count 305 × 109/L (reference range 150–350)
- Orange-colored urine
- Hepatitis C virus (HCV) antibody positive (new finding)
- Human immunodeficiency virus antibody, hepatitis B surface antigen, and antinuclear antibody negative
- Phenytoin and urine drug screen negative
- Aspartate aminotransferase 70 U/L (reference range 5–34)
- Alanine aminotransferase 73 U/L (reference range 0–55)
- Prothrombin time 10.8 seconds (reference range 8.3–13.0), international normalized ratio 0.98 (reference range 0.8–1.2)
- Iron studies within normal limits.
The patient is admitted to the hospital and is started on cefazolin and clindamycin. Urine is collected for a porphyrin screen, and punch-biopsy samples from the forearms are sent for study. Ultrasonography shows splenomegaly, as well as increased echogenicity of the liver without structural abnormalities. Blood and urine cultures, drawn upon admission, are negative by discharge.
Pathologic study of the punch-biopsy specimens (Figure 2) shows the formation of subepidermal vesicles with extensive reticular and dermal fibrosis.

DIAGNOSIS: PORPHYRIA CUTANEA TARDA
Because of the patient’s history, examination, and pathology results, he was preliminarily diagnosed with porphyria cutanea tarda (PCT).1,2 The diagnosis was confirmed after he was discharged when his urine uroporphyrin level was found to be 157.5 μmol/mol of creatinine (reference range < 4) and his urine heptacarboxylporphyrin level was 118.0 μmol/mol of creatinine (reference range < 2).
This patient’s clinical presentation is classic for sporadic (ie, type 1) PCT. Sporadic PCT is an acquired deficiency of uroporphyrinogen decarboxylase, an enzyme that catalyzes the fifth step in heme metabolism.3 The deficiency of this enzyme is exclusively hepatic and is strongly associated with chronic hepatitis C infection. Mutations of the hemochromatosis gene (HFE), human immunodeficiency virus infection, alcohol use, and smoking are also risk factors.4 The prevalence in the United States is about 1:25,000; nearly 80% of cases are sporadic (type 1), and 20% are familial (type 2).5
Manifestations of PCT include photosensitive dermatitis, facial hypertrichosis, and orange urine.3 The photosensitivity dermatitis heals slowly and leads to sclerosis and hyperpigmentation.
Repeated phlebotomy is the first-line treatment, and hydroxychloroquine (Plaquenil) is the second-line treatment.6 Patients with PCT and hepatitis C should be considered for antiviral therapy according to standard guidelines. Treatment of hepatitis C may reduce the symptoms of PCT, even without a sustained viral response. However, not enough evidence exists to make treatment recommendations for this group.7
Because we were uncertain that the patient would return for follow-up, we did not start phlebotomy or treatment for hepatitis C. However, we did prescribe hydroxychloroquine 100 mg three times a week and instructed him to cover his skin when outside and to use effective sunblock. An outpatient visit was scheduled prior to discharge. Unfortunately, the patient was lost to follow-up.
Acknowledgment: The authors would like to personally thank Dr. Karen DeSouza from the University of Tennessee, Graduate School of Medicine, Department of Pathology, for her clinical expertise and kind advice.
- The University of Iowa, Department of Pathology, Laboratory Services Handbook. Porphyrins & Porphobilinogen, Urine (24 hr or random). www.healthcare.uiowa.edu/path_handbook/handbook/test2893.html. Accessed August 8, 2014.
- Maynard B, Peters MS. Histologic and immunofluorescence study of cutaneous porphyrias. J Cutan Pathol 1992; 19:40–47.
- Thunell S, Harper P. Porphyrins, porphyrin metabolism, porphyrias. III. Diagnosis, care and monitoring in porphyria cutanea tarda—suggestions for a handling programme. Scand J Clin Lab Invest 2000; 60:561–579.
- Lambrecht RW, Thapar M, Bonkovsky HL. Genetic aspects of porphyria cutanea tarda. Semin Liver Dis 2007; 27:99–108.
- Kushner JP, Barbuto AJ, Lee GR. An inherited enzymatic defect in porphyria cutanea tarda: decreased uroporphyrinogen decarboxylase activity. J Clin Invest 1976; 58:1089–1097.
- Singal AK, Kormos-Hallberg C, Lee C, et al. Low-dose hydroxychloroquine is as effective as phlebotomy in treatment of patients with porphyria cutanea tarda. Clin Gastroenterol Hepatol 2012; 10:1402–1409.
- Ryan Caballes F, Sendi H, Bonkovsky HL. Hepatitis C, porphyria cutanea tarda and liver iron: an update. Liver Int 2012; 32:880–893.
- The University of Iowa, Department of Pathology, Laboratory Services Handbook. Porphyrins & Porphobilinogen, Urine (24 hr or random). www.healthcare.uiowa.edu/path_handbook/handbook/test2893.html. Accessed August 8, 2014.
- Maynard B, Peters MS. Histologic and immunofluorescence study of cutaneous porphyrias. J Cutan Pathol 1992; 19:40–47.
- Thunell S, Harper P. Porphyrins, porphyrin metabolism, porphyrias. III. Diagnosis, care and monitoring in porphyria cutanea tarda—suggestions for a handling programme. Scand J Clin Lab Invest 2000; 60:561–579.
- Lambrecht RW, Thapar M, Bonkovsky HL. Genetic aspects of porphyria cutanea tarda. Semin Liver Dis 2007; 27:99–108.
- Kushner JP, Barbuto AJ, Lee GR. An inherited enzymatic defect in porphyria cutanea tarda: decreased uroporphyrinogen decarboxylase activity. J Clin Invest 1976; 58:1089–1097.
- Singal AK, Kormos-Hallberg C, Lee C, et al. Low-dose hydroxychloroquine is as effective as phlebotomy in treatment of patients with porphyria cutanea tarda. Clin Gastroenterol Hepatol 2012; 10:1402–1409.
- Ryan Caballes F, Sendi H, Bonkovsky HL. Hepatitis C, porphyria cutanea tarda and liver iron: an update. Liver Int 2012; 32:880–893.
Hypertrophic cardiomyopathy apical variant
He had had an isolated syncopal episode while intensely training a year ago, but his medical history was otherwise unremarkable.
On examination, he appeared fit. His vital signs were normal. The apical pulse was sustained on palpation and was not displaced. Auscultation revealed an S4 heart sound.
HYPERTROPHIC CARDIOMYOPATHY: THE APICAL VARIANT
In typical hypertrophic cardiomyopathy, the left ventricle, especially the interventricular septum, is thickened, but the left ventricular chamber size is normal or small. In severe cases, the left ventricular outflow tract can become very narrowed, resulting in accelerated blood flow, which may further increase in the presence of hypovolemia, peripheral vasodilation, and increased cardiac contractility. The Venturi effect thus created may entrain a typically malformed anterior mitral valve leaflet toward the aortic valve (systolic anterior motion), causing mitral insufficiency and exacerbating obstruction of the left ventricular outflow tract. Systolic anterior motion may play an important role in exercise-induced syncope and sudden death in young people with hypertrophic cardiomyopathy.4
In the apical variant, hypertrophy is confined to the left ventricular apex.1–3 There is no dynamic outflow tract obstruction. Still, unexplained syncope has been reported, and recent data challenge the conventional wisdom that the apical variant of hypertrophic cardiomyopathy has a benign prognosis.2 In patients without a history of recurrent syncope, chest pain, or heart failure, perioperative risk is probably not significantly increased. The differential diagnosis includes myocardial ischemia or infarction, electrolyte disturbances, effects of drugs (eg, digoxin), and subarachnoid hemorrhage.1–3,5 Plain or contrast-enhanced echocardiography or cardiac magnetic resonance imaging, or both, can help confirm the diagnosis. Long-term management should be guided by the patient’s symptoms.
- Eriksson MJ, Sonnenberg B, Woo A, et al. Long-term outcome in patients with apical hypertrophic cardiomyopathy. J Am Coll Cardiol 2002; 39:638–645.
- Kasirye Y, Manne JR, Epperla N, Bapani S, Garcia-Montilla R. Apical hypertrophic cardiomyopathy presenting as recurrent unexplained syncope. Clin Med Res 2012; 10:26–31.
- Lee CH, Liu PY, Lin LJ, Chen JH, Tsai LM. Clinical features and outcome of patients with apical hypertrophic cardiomyopathy in Taiwan. Cardiology 2006; 106:29–35.
- Nishimura RA, Holmes DR Clinical practice. Hypertrophic obstructive cardiomyopathy. N Engl J Med 2004; 350:1320–1327.
- Lin CS, Chen CH, Ding PY. Apical hypertrophic cardiomyopathy mimicking acute myocardial infarction. Int J Cardiol 1998; 64:305–307.
He had had an isolated syncopal episode while intensely training a year ago, but his medical history was otherwise unremarkable.
On examination, he appeared fit. His vital signs were normal. The apical pulse was sustained on palpation and was not displaced. Auscultation revealed an S4 heart sound.
HYPERTROPHIC CARDIOMYOPATHY: THE APICAL VARIANT
In typical hypertrophic cardiomyopathy, the left ventricle, especially the interventricular septum, is thickened, but the left ventricular chamber size is normal or small. In severe cases, the left ventricular outflow tract can become very narrowed, resulting in accelerated blood flow, which may further increase in the presence of hypovolemia, peripheral vasodilation, and increased cardiac contractility. The Venturi effect thus created may entrain a typically malformed anterior mitral valve leaflet toward the aortic valve (systolic anterior motion), causing mitral insufficiency and exacerbating obstruction of the left ventricular outflow tract. Systolic anterior motion may play an important role in exercise-induced syncope and sudden death in young people with hypertrophic cardiomyopathy.4
In the apical variant, hypertrophy is confined to the left ventricular apex.1–3 There is no dynamic outflow tract obstruction. Still, unexplained syncope has been reported, and recent data challenge the conventional wisdom that the apical variant of hypertrophic cardiomyopathy has a benign prognosis.2 In patients without a history of recurrent syncope, chest pain, or heart failure, perioperative risk is probably not significantly increased. The differential diagnosis includes myocardial ischemia or infarction, electrolyte disturbances, effects of drugs (eg, digoxin), and subarachnoid hemorrhage.1–3,5 Plain or contrast-enhanced echocardiography or cardiac magnetic resonance imaging, or both, can help confirm the diagnosis. Long-term management should be guided by the patient’s symptoms.
He had had an isolated syncopal episode while intensely training a year ago, but his medical history was otherwise unremarkable.
On examination, he appeared fit. His vital signs were normal. The apical pulse was sustained on palpation and was not displaced. Auscultation revealed an S4 heart sound.
HYPERTROPHIC CARDIOMYOPATHY: THE APICAL VARIANT
In typical hypertrophic cardiomyopathy, the left ventricle, especially the interventricular septum, is thickened, but the left ventricular chamber size is normal or small. In severe cases, the left ventricular outflow tract can become very narrowed, resulting in accelerated blood flow, which may further increase in the presence of hypovolemia, peripheral vasodilation, and increased cardiac contractility. The Venturi effect thus created may entrain a typically malformed anterior mitral valve leaflet toward the aortic valve (systolic anterior motion), causing mitral insufficiency and exacerbating obstruction of the left ventricular outflow tract. Systolic anterior motion may play an important role in exercise-induced syncope and sudden death in young people with hypertrophic cardiomyopathy.4
In the apical variant, hypertrophy is confined to the left ventricular apex.1–3 There is no dynamic outflow tract obstruction. Still, unexplained syncope has been reported, and recent data challenge the conventional wisdom that the apical variant of hypertrophic cardiomyopathy has a benign prognosis.2 In patients without a history of recurrent syncope, chest pain, or heart failure, perioperative risk is probably not significantly increased. The differential diagnosis includes myocardial ischemia or infarction, electrolyte disturbances, effects of drugs (eg, digoxin), and subarachnoid hemorrhage.1–3,5 Plain or contrast-enhanced echocardiography or cardiac magnetic resonance imaging, or both, can help confirm the diagnosis. Long-term management should be guided by the patient’s symptoms.
- Eriksson MJ, Sonnenberg B, Woo A, et al. Long-term outcome in patients with apical hypertrophic cardiomyopathy. J Am Coll Cardiol 2002; 39:638–645.
- Kasirye Y, Manne JR, Epperla N, Bapani S, Garcia-Montilla R. Apical hypertrophic cardiomyopathy presenting as recurrent unexplained syncope. Clin Med Res 2012; 10:26–31.
- Lee CH, Liu PY, Lin LJ, Chen JH, Tsai LM. Clinical features and outcome of patients with apical hypertrophic cardiomyopathy in Taiwan. Cardiology 2006; 106:29–35.
- Nishimura RA, Holmes DR Clinical practice. Hypertrophic obstructive cardiomyopathy. N Engl J Med 2004; 350:1320–1327.
- Lin CS, Chen CH, Ding PY. Apical hypertrophic cardiomyopathy mimicking acute myocardial infarction. Int J Cardiol 1998; 64:305–307.
- Eriksson MJ, Sonnenberg B, Woo A, et al. Long-term outcome in patients with apical hypertrophic cardiomyopathy. J Am Coll Cardiol 2002; 39:638–645.
- Kasirye Y, Manne JR, Epperla N, Bapani S, Garcia-Montilla R. Apical hypertrophic cardiomyopathy presenting as recurrent unexplained syncope. Clin Med Res 2012; 10:26–31.
- Lee CH, Liu PY, Lin LJ, Chen JH, Tsai LM. Clinical features and outcome of patients with apical hypertrophic cardiomyopathy in Taiwan. Cardiology 2006; 106:29–35.
- Nishimura RA, Holmes DR Clinical practice. Hypertrophic obstructive cardiomyopathy. N Engl J Med 2004; 350:1320–1327.
- Lin CS, Chen CH, Ding PY. Apical hypertrophic cardiomyopathy mimicking acute myocardial infarction. Int J Cardiol 1998; 64:305–307.
Polycystic kidney disease: Molecular understanding dictating management
Dr. Braun is an iconic figure in Cleveland Clinic medicine. He is the consummate internist, nephrologist, and transplantation physician, but he is also a critical thinker. He strives to understand (and explain) what underpins our clinical observations and therapeutic decisions. He asks the “why” questions. As he ticked through the manifestations of PKD and the diagnostic dilemmas that arise in taking care of these patients, and then transitioned into explaining the interesting though incomplete current molecular understanding of this relatively prevalent genetic disorder, I heard many of the same questions I had asked myself 30 years ago. But this time I was getting some answers.
How can one be certain a cyst is infected? How do these cysts form and expand without apparent communication with the tubular lumens? (Intracystic bleeding and infection may not be reflected in the urinalysis, although the organism isolated from infected cysts is frequently Escherichia coli.) If renal cysts are formed from tubular epithelial cells that are preprogrammed to self-organize into lumen-like structures, how does the same genetic defect predispose to cyst formation in organs such as the liver, or to aneurysms in blood vessels in the brain? Why does the disease take so long to express itself, and why is its expression so variable?
The patient did well during his hospital stay 30 years ago. As I recall, he had staphylococcal bacteremia with an infected cyst. We discussed the clinical scenario but had no suggestions as to how to prevent the growth of what we now know are about 60 subclinical cysts for every one that we recognize. And we certainly didn’t discuss the idea that the disease process may be partially driven by dysfunctional nonmotile cilia that should respond to urine flow by appropriately directing regeneration and proliferation of renal tubular cells.
I love getting answers to questions that I didn’t know enough to ask.
Dr. Braun is an iconic figure in Cleveland Clinic medicine. He is the consummate internist, nephrologist, and transplantation physician, but he is also a critical thinker. He strives to understand (and explain) what underpins our clinical observations and therapeutic decisions. He asks the “why” questions. As he ticked through the manifestations of PKD and the diagnostic dilemmas that arise in taking care of these patients, and then transitioned into explaining the interesting though incomplete current molecular understanding of this relatively prevalent genetic disorder, I heard many of the same questions I had asked myself 30 years ago. But this time I was getting some answers.
How can one be certain a cyst is infected? How do these cysts form and expand without apparent communication with the tubular lumens? (Intracystic bleeding and infection may not be reflected in the urinalysis, although the organism isolated from infected cysts is frequently Escherichia coli.) If renal cysts are formed from tubular epithelial cells that are preprogrammed to self-organize into lumen-like structures, how does the same genetic defect predispose to cyst formation in organs such as the liver, or to aneurysms in blood vessels in the brain? Why does the disease take so long to express itself, and why is its expression so variable?
The patient did well during his hospital stay 30 years ago. As I recall, he had staphylococcal bacteremia with an infected cyst. We discussed the clinical scenario but had no suggestions as to how to prevent the growth of what we now know are about 60 subclinical cysts for every one that we recognize. And we certainly didn’t discuss the idea that the disease process may be partially driven by dysfunctional nonmotile cilia that should respond to urine flow by appropriately directing regeneration and proliferation of renal tubular cells.
I love getting answers to questions that I didn’t know enough to ask.
Dr. Braun is an iconic figure in Cleveland Clinic medicine. He is the consummate internist, nephrologist, and transplantation physician, but he is also a critical thinker. He strives to understand (and explain) what underpins our clinical observations and therapeutic decisions. He asks the “why” questions. As he ticked through the manifestations of PKD and the diagnostic dilemmas that arise in taking care of these patients, and then transitioned into explaining the interesting though incomplete current molecular understanding of this relatively prevalent genetic disorder, I heard many of the same questions I had asked myself 30 years ago. But this time I was getting some answers.
How can one be certain a cyst is infected? How do these cysts form and expand without apparent communication with the tubular lumens? (Intracystic bleeding and infection may not be reflected in the urinalysis, although the organism isolated from infected cysts is frequently Escherichia coli.) If renal cysts are formed from tubular epithelial cells that are preprogrammed to self-organize into lumen-like structures, how does the same genetic defect predispose to cyst formation in organs such as the liver, or to aneurysms in blood vessels in the brain? Why does the disease take so long to express itself, and why is its expression so variable?
The patient did well during his hospital stay 30 years ago. As I recall, he had staphylococcal bacteremia with an infected cyst. We discussed the clinical scenario but had no suggestions as to how to prevent the growth of what we now know are about 60 subclinical cysts for every one that we recognize. And we certainly didn’t discuss the idea that the disease process may be partially driven by dysfunctional nonmotile cilia that should respond to urine flow by appropriately directing regeneration and proliferation of renal tubular cells.
I love getting answers to questions that I didn’t know enough to ask.
Advances in autosomal dominant polycystic kidney disease—2014 and beyond
Autosomal dominant polycystic kidney disease (ADPKD) is the most common inherited renal disease, has an estimated prevalence of 1:400 to 1:1,000 live births in the United States, and occurs worldwide.1,2 There are about 700,000 people living with it in the United States, and about 6,000 new cases arise annually. It accounts for nearly 5% of all patients with end-stage renal disease in the United States.3
This paper will offer an overview of the pathogenesis of renal cysts, review some of the clinical aspects of ADPKD including diagnosis and management of complications, and discuss recent drug trials and current management.
TWO TYPES—PKD1 IS MORE COMMON AND PROGRESSES MORE RAPIDLY
Two major forms of ADPKD are recognized and can usually be determined by genetic testing: PKD1, accounting for about 85% of cases, and PKD2, accounting for 15%.
The gene locus for PKD1 is on the short arm of the 16th chromosome (16p13.3), and its glycoprotein gene product is polycystin 1 (PC1), a large molecule with 4,303 amino acids.2 PC1 has a long N-terminal extracellular tail that can function as a mechanosensor. Disease progression is much faster with PKD1, and end-stage renal disease usually occurs before age 56.4
In PKD2, the gene locus is on the long arm of the fourth chromosome (4q21–23), and has a smaller glycoprotein gene product, polycystin 2 (PC2), that plays a role in calcium transport. The disease course of PKD2 tends to be slower. End-stage renal disease might not develop in the patient’s lifetime, since it typically develops when the patient is more than 70 years old.4
Although the growth rate of renal cysts is similar between the two types, patients with PKD1 develop about twice as many cysts as those with PDK2, and their cyst development starts at a younger age.5
Typically, patients have a clear phenotype and a positive family history, but in about 10% of possible ADPKD cases, there is no family history of ADPKD. Genetic variations such as incompletely penetrant PKD1 alleles,6 hypomorphic alleles,7 and trans-heterozygous mutations8 account for at least some of these cases.
IMAGING CRITERIA HAVE BROADENED
Ultrasonographic criteria for the diagnosis of ADPKD that were published in 1994 were based on patients who had a family history of PKD1.9 The criteria have since been modified (the “unified criteria”) to include patients with a family history of PKD2 who begin cyst development at a later age and with lower numbers.10 For patients ages 30 to 39, a previously difficult diagnostic group, the criterion for the minimum number of cysts visible on ultrasonography changed from four to three, improving the sensitivity of detecting disease from approximately 76% to approximately 95% (Table 1).9,10 It is important to note that these criteria apply only to patients “at risk,” ie, with a positive family history of ADPKD.
Computed tomography (CT) and magnetic resonance imaging (MRI) classically show bilaterally enlarged multicystic kidneys, though variations can be seen.
DISEASE CAN PRESENT IN MYRIAD WAYS
Although cystic kidney disease is the basic underlying problem, undiagnosed patients may present with a variety of symptoms caused by other manifestations of ADPKD (Table 2).
Hypertension is the most common presentation, occurring in about 50% of patients ages 20 to 34, and essentially 100% of those with end-stage renal disease.11 It is associated with up-regulation of the renin-angiotensin-aldosterone system.
Pain is typically located in the abdomen, flank, or back and can occur in a localized or diffuse manner. Early abdominal distress is often simply described as “fullness.” Localized pain is usually caused by bleeding into or rupture of a cyst, renal stones, or infection.12 Because renal cysts are noncommunicating, bleeding can occur into a cyst and cause pain without gross hematuria. Compression by greatly enlarged kidneys, liver, or both can cause a variety of gastrointestinal symptoms such as reflux esophagitis and varying degrees of constipation. Diffuse pain is often musculoskeletal and related to exaggerated lordosis from increasing abdominal size due to enlarging cystic kidneys and sometimes liver.12 In carefully selected cases, cyst aspiration may be helpful.11
Although renal carcinomas are rare and not more frequent than in the general population, they can occur at an earlier age and with constitutional symptoms.11
Urinary tract infections are increased in frequency. A patient may have a simple urinary tract infection that is cured with the appropriate antibiotic. However, a urinary tract infection repeatedly recurring with the same organism is a strong clue that an infected cyst is the source and requires more intensive treatment with the appropriate cyst-penetrating antibiotic. On the other hand, because cysts are noncommunicating, an infected cyst might be present despite a negative urine culture.
Identifying infected cysts can be a challenge with conventional imaging techniques, but combined positron emission tomography and CT (PET/CT) can be a valuable though expensive diagnostic tool to identify an infected kidney or liver cyst, or to identify an unsuspected source of the pain and infection.13
Jouret et al13 evaluated 27 PET/CT scans performed in 24 patients with ADPKD and suspicion of an abdominal infection. Patients were deemed to have probable cyst infection if they met all of the following criteria: temperature more than 38°C for longer than 3 days, loin or liver tenderness, plasma C-reactive protein level greater than 5 mg/dL, and no evidence of intracystic bleeding on CT. Patients with only two or three of these criteria were classified as having fever of unknown origin. Diagnosis of cyst infection was confirmed by cyst fluid analysis.
PET/CT identified a kidney or liver cyst infection in 85% of 13 infectious events in 11 patients who met all the criteria for probable cyst infection; CT alone contributed to the diagnosis in only one patient.13 In those with fever of unknown origin, PET/CT identified a source of infection in 64% of 14 events in 13 patients: two infected renal cysts, as well as one patient each with other infections that would be difficult to diagnose clinically, ie, small bowel diverticulitis, psoas abscess, diverticulitis of the right colon, pyelonephritis in a transplanted kidney, infected abdominal aortic aneurysm, prostatitis, colitis, and Helicobacter pylori gastritis. Results of PET/CT were negative in five patients with intracystic bleeding.
Kidney stones occur in 20% to 36% of patients.11,14 Uric acid stones occur at almost the same frequency as calcium oxalate stones.
Chronic kidney disease not previously diagnosed may be the presenting condition in a small percentage of patients, sometimes those in whom much earlier hypertension was not fully evaluated. ADPKD is typically not associated with significant proteinuria (eg, nephrotic range), and the presence of heavy proteinuria usually indicates the presence of a superimposed primary glomerulopathy.15
Cysts in other locations. By MRI, liver cysts are present in 58% of patients ages 15 to 24, rising to 94% in those ages 35 to 46.11 Because liver cysts are estrogen-dependent, they are more prominent in women. A small percentage of patients develop cysts in the pancreas (5%), arachnoid membranes (8%), and seminal vesicles (40% of men with ADPKD).11
Cardiovascular abnormalities occur in almost one-third of patients with ADPKD, usually as mitral and aortic valve abnormalities.16 Aneurysms of the aortic root and abdominal aorta can also occur, in addition to intracranial aneurysms (see below).17
Intracranial aneurysms are not uncommon, and size usually determines their risk.
Intracranial aneurysms are strongly influenced by family history: 16% of ADPKD patients with a family history of intracranial aneurysm also develop them, compared with 5% to 6% of patients with no family history.11 The anterior cerebral circulation is involved in about 80% of cases. A sentinel or sudden “thunderclap” headache is a classic presentation that may precede full-blown rupture in about 17% of cases.18 Patients who rupture an intracranial aneurysm have a mean age of 39, usually have normal renal function, and can be normotensive.11
For patients with no history of subarachnoid hemorrhage, the 5-year cumulative rupture rates for patients with aneurysms located in the internal carotid artery, anterior communicating or anterior cerebral artery, or middle cerebral artery were 0% for aneurysms less than 7 mm, 2.6% for those 7 to 12 mm, 14.5% for those 13 to 24 mm, and 40% for those 25 mm or larger, with higher rates for the same sizes in the posterior circulation.11
In patients without symptoms, size is correlated with risk of rupture: less than 4 mm is usually associated with very low risk, 4 to less than 7 mm with moderate risk, and 7 mm or more with increasing risk. An aneurysm larger than 10 mm is associated with roughly a 1% risk of rupture per year.19
Irazabal et al20 retrospectively studied 407 patients with ADPKD who were screened for intracranial aneurysm. Saccular aneurysms were detected in 45 patients; most were small (median diameter 3.5 mm). During cumulative imaging follow-up of 243 years, only one new intracranial aneurysm was detected (increasing from 2 to 4.4 mm over 144 months) and two previously identified aneurysms grew (one increasing 4.5 to 5.9 mm over 69 months and the other 4.7 to 6.2 mm over 184 months). No change occurred in 28 patients. Seven patients were lost to follow-up, however. During cumulative clinical follow-up of 316 years, no aneurysm ruptured. Two patients were lost to follow-up, three had surgical clipping, and five died of unrelated causes. The authors concluded that presymptomatic intracranial aneurysms are usually small, and that growth and rupture risks are no higher than for unruptured intracranial aneurysms in the general population. A 2014 study also suggests a conservative approach for managing intracranial aneurysm in the general population.21
In asymptomatic ADPKD patients, it is reasonable to reserve screening for those with a positive family history of intracranial aneurysm or subarachnoid hemorrhage, those with a previous ruptured aneurysm, those in high-risk professions (eg, pilots), and for patients prior to anticoagulant therapy or major surgery possibly associated with hemodynamic instability.11,22 Certain extremely anxious patients might also need to be studied. Screening can be performed with magnetic resonance angiography without gadolinium contrast. It is prudent to have patients with an intracranial aneurysm thoroughly evaluated by an experienced neurosurgeon with continued follow-up.
PROGRESSION OF ADPKD
The Consortium for Radiologic Imaging Studies of Polycystic Kidney Disease (CRISP) study23 evaluated 241 patients with ADPKD (ages 15 to 46) by measuring the annual rate of change in total kidney volume, total cyst volume, and iothalamate glomerular filtration rate (GFR) over 3 years. The annual increase in total kidney volume averaged 5.3%,23 though the reported range with various imaging techniques is from 4% to 12.8% in adults.24 This study focused on macrocystic disease, ie, cysts that are visible by MRI and measurably increase total kidney volume. Although larger total kidney volume at baseline generally predicted a more rapid decline in GFR, there were wide and overlapping variations in yearly GFR declines within and among different total-kidney-volume groups.23
SPECIAL CLINICAL PROBLEMS IN ADPKD
Case 1: A man with ADPKD develops new and increasing proteinuria
A 55-year-old man with ADPKD and stage 3 chronic kidney disease developed new and increasing proteinuria, rising to 5,500 mg per 24 hours. What is the most likely explanation?
- Rapidly progressive renal failure with increasing proteinuria in ADPKD
- Bilateral renal vein thromboses because of cyst compression
- Malignant hypertension with bilateral renal artery compression
- Superimposed primary glomerulopathy
- Multiple infected renal cysts with pyonephrosis
Answer: Superimposed primary glomerulopathy.
ADPKD (similar to uncomplicated obstructive uropathy, pyelonephritis, main renal artery disease, and often cases of interstitial nephritis without secondary glomerular changes) typically does not result in nephrotic-range proteinuria. A superimposed primary glomerulopathy, focal segmental glomerulosclerosis, was the biopsy-proved diagnosis.
At least 21 cases have been reported of AD-PKD with nephrotic-range proteinuria and a renal biopsy showing a primary glomerulopathy, including focal segmental glomerulosclerosis (5 cases), minimal-change disease (5), membranous nephropathy (3), IgA nephropathy (2), and one each of crescentic glomerulonephropathy, diabetic nephropathy, membranoproliferative glomerulonephritis, postinfectious glomerulonephropathy, amyloid glomerulopathy, and mesangioproliferative glomerulopathy.15 Treatment was directed at the primary glomerulopathy, and the outcomes corresponded to the primary diagnosis (eg, with appropriate treatment, three of the five patients with focal segmental glomerulosclerosis progressed to end-stage renal disease, all of the patients with minimal-change disease went into remission, and one of the two cases with IgA nephropathy improved).15
Case 2: A woman with ADPKD and advanced renal failure develops shortness of breath
A 47-year-old woman with very large polycystic kidneys (total kidney volume 7,500 mL; normal range for a single kidney approximately 136–295 mL, mean 196)25 and estimated GFR of 25 mL/min developed new-onset shortness of breath while climbing steps and later even when making a bed. She had no chest pain, cough, or edema. She was sent directly to the emergency department and was admitted and treated; her condition improved, and she was discharged after 6 days. What did she have?
- Presentation of rare cystic pulmonary disease in ADPKD
- Onset of pneumonia with early bacteremia
- Progressive reduction in ventilatory capacity from massive polycystic kidneys and liver elevating both sides of the diaphragm
- Pulmonary emboli from an iliac vein or inferior vena cava source
- Progressive anemia accompanying rapidly worsening stage 4 chronic kidney disease
Answer: She had pulmonary emboli from an iliac vein (right) or inferior vena cava source.
Pulmonary emboli in ADPKD can be caused by thrombi in the inferior vena cava or the iliac or femoral vein because of compression by a massive right polycystic kidney. Four cases were reported at Mayo Clinic,26 three diagnosed by MRI and one with CT. One additional case occurred at Cleveland Clinic. All patients survived after treatment with anticoagulation therapy; early nephrectomy was required in two cases.
Interestingly, following kidney transplantation, the patients at greatest risk for pulmonary emboli are those with ADPKD as their original disease.27
RENAL CYSTS RESULT FROM COMBINED MUTATIONS, INJURY
The germline ADPKD mutation that occurs in one allele of all renal tubular epithelial cells is necessary but not sufficient for cystogenesis.28 One or more additional somatic mutations of the normal allele—the “second hit”—also develop within individual tubular epithelial cells.28,29 These epithelial cells undergo clonal proliferation, resulting in tubular dilatation and cyst formation. Monoclonality of cells in cysts has been documented.
Ischemia-reperfusion injury can be viewed as a “third hit.”30 In PKD1 knockout mice, which at 5 weeks of age normally develop only mild cystic kidney disease, the superimposition of unilateral ischemia-reperfusion injury at 8 weeks caused widespread and rapid cyst formation. It is believed that acute renal injury reactivates developmental signaling pathways within 48 hours that trigger epithelial cell proliferation and then cyst development detectable by MRI 2 weeks later. Although this phenomenon has not been documented in humans, it is a cautionary tale.
CYSTOGENESIS INVOLVES MULTIPLE PATHWAYS
A comprehensive description of pathways leading to renal cyst formation is beyond the scope of this article, and the reader is referred to much more detailed and extensive reviews.2,31 Disturbances in at least three major interconnected pathways promote cystogenesis in renal tubular epithelial cells:
- Normal calcium transport into the endoplasmic reticulum is disrupted by abnormal polycystins on the surface of the primary cilium
- Vasopressin and other stimuli increase the production of cyclic adenosine monophosphate (cAMP)
- The mammalian target of rapamycin (mTOR) proliferative pathway is up-regulated.
DISRUPTION OF CALCIUM TRANSPORT IN THE PRIMARY CILIUM
Primary cilia are nonmotile cellular organelles of varying size, from about 0.25 μm up to about 1 μm.32 Each primary cilium has nine peripheral pairs of microtubules but lacks a centrally located pair that is present in motile cilia. Primary cilia are ubiquitous and have been highly conserved throughout evolution. A single cilium is present on almost all vertebral cells.33
Cilial defects have been identified in autosomal dominant as well as recessive diseases and are known as ciliopathies.33 Although rare in humans, they can affect a broad spectrum of organs other than the kidney, including the eye, liver, and brain.33
Urine flow in a renal tubule is believed to exert mechanical stimulation on the extracellular flagellum-like N-terminal tail of PC1 that extends from a primary cilium into the urinary space. PC1 in concert with PC2 opens PC2 calcium channels, allowing calcium ions to flow down the microtubules to ryanodine receptors and the basal body.32,33 This leads to local release of calcium ions that regulate cell proliferation.32,34 However, in ADPKD kidneys, PC1 and PC2 molecules are sparse or mutated, resulting in defective calcium transport, increased and unregulated tubular epithelial cell proliferation, and cyst formation.
In a totally different clinical setting, biopsies of human renal transplants that sustained acute tubular necrosis during transplantation reveal that a cilium dramatically elongates in response to injury,35 possibly as a compensatory mechanism to maintain calcium transport in the presence of meager urine flow and to restore the proliferation of tubular epithelial cells in a regulated repair process.
THE ROLE OF VASOPRESSIN AND ACTIVATION OF cAMP
In classic experiments, Wang et al36 cross-bred rats having genetically inherited polycystic kidney disease (actually, autosomal recessive polycystic kidney disease) with Brattleboro rats that completely lack vasopressin. At 10 and 20 weeks of age, the offspring had virtually complete inhibition of cystogenesis because of the absence of vasopressin. However, when vasopressin was restored by exogenous administration continuously for 8 weeks, the animals formed massive renal cysts.
Vasopressin activates cAMP, which then functions as a second messenger in cell signaling. cAMP increases the activation of the protein kinase A (PKA) pathway, which in turn increases downstream activity of the B-raf/ERK pathway. Up-regulation of cAMP and PKA appears to perpetuate activation of canonical Wnt signaling, down-regulate non-canonical Wnt/planar cell polarity signaling, and lead to loss of tubular diameter control, resulting in cyst formation.31 Normally, cAMP is degraded by phosphodiesterase. However, because of the primary cilium calcium transport defect in ADPKD, phosphodiesterase is reduced and cAMP persists.37 In conjunction with the defective primary cilial calcium transport, cAMP exerts a proliferative effect on renal tubular epithelial cells that is opposite to its effect in normal kidneys.31,32 cAMP also up-regulates the cystic fibrosis transmembrane conductance regulator (CFTR) that promotes chloride ion transport. Sodium ions follow the chloride ions, leading to fluid accumulation and cyst enlargement.31
Inhibiting vasopressin by increasing water intake
A simple key mechanism for limiting vasopressin secretion is by sufficient water ingestion. Nagao et al38 found that rats with polycystic kidney disease given water with 5% glucose (resulting in 3.5-fold increased fluid intake compared with rats given tap water) had a 68% reduction in urinary vasopressin and a urine osmolality less than 290 mOsm/kg. The high-water-intake rats had dramatically reduced cystic areas in the kidney and a 28% reduction of kidney-to-body weight ratio vs controls.
In an obvious oversimplification, these findings raised the question of whether a sufficient increase in water intake could be an effective therapy for polycystic kidney disease.39 A pilot clinical study evaluated changes in urine osmolality in eight patients with ADPKD who had normal renal function.40 At baseline, 24-hour urine osmolality was typically elevated to approximately 753 mOsm/kg compared to the plasma at 285 mOsm/kg, indicating that antidiuresis is the usual state. During the 2-week study, urine volume and osmolality were measured, and additional water intake was adjusted in order to achieve a urine osmolality goal of 285 ± 45 mOsm/kg. These adjustments resulted in water intake that appeared to be in the range of 2,400 to 3,000 mL per 24 hours. The major limitations of the study were that it was very short term, and there was no opportunity to measure changes in total kidney volume or estimated GFR.
In a recent preliminary report from Japan, high water intake (2,500–3,000 mL daily) in 18 ADPKD patients was compared over 12 months with ad libitum water intake in 14 ADPKD controls (clinicaltrials.gov NCT 01348505). There was no statistically significant change in total kidney volume or cystatin-estimated GFR in those on high water intake, but serious defects in study design (patients in the high water intake group were allowed to decrease their intake if it was causing them difficulty, and patients in the ad libitum water intake group had no measurement of their actual water intake) prevent any conclusions because there was no evidence that the groups were different from one another with respect to the key element of the study, namely, water intake.
Blocking the vasopressin receptor slows disease progression
Using another approach, Gattone et al41 inhibited the effect of vasopressin by blocking the vasopressin 2 receptor (V2R) in mouse and rat models of polycystic kidney disease, using an experimental drug, OPC31260. The drug halted disease progression and, in one situation, appeared to cause regression of established disease. As noted by Torres and Harris,31 even though both increased water intake and V2R antagonists decrease cAMP in the distal tubules and collecting ducts, circulating levels of vasopressin are decreased by increased water intake but increased by V2R antagonists.
After these remarkable results in animal models, clinical trials of the V2R antagonist tolvaptan were conducted in patients with ADPKD. In the Tolvaptan Efficacy and Safety in Management of Autosomal Dominant Polycystic Kidney Disease and Its Outcomes 3:4 study,42 1,445 adults (ages 18 to 50) with ADPKD in 133 centers worldwide were randomized to receive either tolvaptan or placebo for 3 years. Key inclusion criteria included good renal function (estimated GFR ≥ 60 mL/min) and total kidney volume of at least 750 mL (mean 1,700 mL) as measured by MRI. Tolvaptan was titrated to the highest tolerated twice-daily dose (average total of 95 mg/day). All patients were advised to maintain good hydration and to avoid thirst by drinking a glass of water after each urination. Unfortunately, neither water intake nor urine output was measured.
The primary end point was the annual rate of change in total kidney volume, with secondary end points of clinical progression (worsening kidney function, pain, hypertension, albuminuria), and rate of decline in kidney function as measured by the slope of the reciprocal of serum creatinine.42
Patients in the tolvaptan arm had a slower annual increase in total kidney volume than controls (2.8% vs 5.5%, respectively, P < .001) and a slower annual decline in renal function (−2.61 vs −3.81 mg/mL−1, respectively, P < .001).42 More participants in the treatment group withdrew than in the placebo group (23% vs 14%, respectively).
Adverse events occurred more frequently with tolvaptan.42 Liver enzyme elevations of greater than three times the upper limit of normal occurred in 4.4% of patients in the treatment group, leading to a drug warning issued in January 2013. As expected, side effects related to diuresis (urinary frequency, nocturia, polyuria, and thirst) were more frequent in the treatment group, occurring in up to 55% of participants.
The authors noted, “Although maintaining hydration helped ensure that the blinding in the study was maintained, the suppression of vasopressin release in the placebo group may have led to an underestimation of the beneficial effect of tolvaptan and may account for the lower rates of kidney growth observed in the placebo group.”42
In 2013, the US Food and Drug Administration (FDA) denied a new drug application for tolvaptan as a treatment for ADPKD.
THE mTOR PATHWAY IS UP-REGULATED
The mTOR pathway that plays a major role in cell growth and proliferation includes interaction of the cytoplasmic tail of polycystin 1 with tuberin.43 Activation products of mTOR, including phospho-S6K, have been found in tubular epithelial cells lining cysts of ADPKD kidneys but not in normal kidneys.43 Mutant mice with polycystic disease had phospho-S6K in tubular epithelial cells of cysts, whereas those treated with the mTOR inhibitor rapamycin did not.43 But subsequent studies have shown that only a low level of mTOR activation is present in 65% to 70% of ADPKD cysts.44
Two major studies of the treatment of ADPKD with rapamycin that were published contemporaneously in 2010 failed to demonstrate any significant benefit with mTOR inhibitor treatment.45,46
Serra et al45 conducted an 18-month, open-label trial of 100 ADPKD patients ages 18 to 40 with an estimated GFR (eGFR) of at least 70 mL/min. Patients were randomized to receive rapamycin, given as sirolimus 2 mg per day, or standard care. The primary end point was the reduction in the growth rate of total kidney volume, measured by MRI. Secondary end points were eGFR and protein excretion (albumin-creatinine ratio). No significant difference was found in total kidney volume, but a nonsignificant stabilization of eGFR was noted.
Walz et al46 in a 2-year, multicenter, double-blind trial, randomized 433 patients (mean age 44; mean eGFR 54.5 mL/min) to treatment with either the short-acting mTOR inhibitor everolimus (2.5 mg twice daily) or placebo. Although patients in the treatment group had less of an increase in total kidney volume (significant at 1 year but not at 2 years), they tended to show a decline in eGFR. But further analysis showed that the only patients who had a reduction in eGFR were males who already had impaired kidney function at baseline.47
In a pilot study, 30 patients with ADPKD (mean age 49) were randomized to one of three therapies:
- Low-dose rapamycin (trough blood level 2–5 ng/mL)
- Standard-dose rapamycin (trough blood level > 5–8 ng/mL)
- Standard care without rapamycin.48
In contrast to other studies, the primary end point was the change in iothalamate GFR at 12 months, with change in total kidney volume being a secondary end point.
At 12 months, with 26 patients completing the study, the low-dose rapamycin group (n = 9) had a significant increase in iothalamate GFR of 7.7 ± 12.5 mL/min/1.73 m2, whereas the standard-dose rapamycin group (n = 8) had a nonsignificant increase of 1.6 ± 12.1 mL/min/1.73 m2, and the no-rapamycin group (n = 9) had a fall in iothalamate GFR of 11.2 ± 9.1 mL/min/1.73 m2 (P = .005 for low-dose vs no rapamycin; P = .07 for standard-dose vs no rapamycin; P = .52 for low-dose vs standard-dose rapamycin; and P = .002 for combined low-dose and standard-dose rapamycin vs no rapamycin.).48 These differences were observed despite there being no significant change in total kidney volume in any of the groups. Patients on low-dose rapamycin had fewer adverse effects than those on standard dose and were more often able to continue therapy for the entire study. This, and the use of iothalamate GFR rather than eGFR to measure GFR, are believed to be the main reasons that low-dose effects were more pronounced than those with standard doses. One may speculate that rapamycin may have its effect on microcysts and cystogenic cells, resulting in stabilization of or improvement in renal function without detectable slowing in total kidney volume enlargement. Mechanisms for this possibility involve new concepts, as discussed below.
NEW CONCEPTS
Specialized cells also promote renal cyst formation
Specialized cells that promote cyst formation have been identified by Karihaloo et al49 in a mouse model of polycystic kidney disease. In this model, alternatively activated macrophages homed to cystic areas and promoted cyst growth. These findings suggested that interrupting the homing and proliferative signals of macrophages could be a therapeutic target for ADPKD. Although rapamycin can suppress macrophage proliferation by macrophage colony-stimulating factor and inhibit macrophage function,50 alternatively activated macrophages have not been specifically studied for rapamycin responsiveness.
More promising is evidence that CD133+ progenitor cells from human ADPKD kidneys—but not from normal human kidneys—form cysts in vitro and in severe combined immunodeficient mouse models.51 Treatment with rapamycin decreased proliferation of the de-differentiated CD133+ cells from ADPKD patients and reduced cystogenesis.51
Visible cysts are the tip of the iceberg
Using ADPKD nephrectomy specimens from eight patients, Grantham et al52 compared cyst counts by MRI and by histology and found that for every renal cyst detected by MRI, about 62 smaller cysts (< 0.9 mm) are present in the kidney. For a typical patient having an average of 587 cysts in both kidneys that are detectable by MRI, this means that more than 36,000 cysts are actually present, and MRI detects less than 2% of the total cysts present.
Although microcysts are too small to contribute much to total kidney volume, they can interfere with kidney function. Microcysts can reduce GFR in two major ways: by compressing microvasculature, tubules, and glomeruli in the cortex; or by blocking the drainage of multiple upstream nephrons when they form in or block medullary collecting ducts.52 Although the growth rates of microcysts less than 1 mm in size have not yet been measured, the adult combined growth rates of the renal cyst component is approximately 12% per year, with yearly individual cyst growth rates up to 71%, and with fetal cyst growth rates even higher for cysts larger than 7.0 mm.53 Before and during an accelerated growth period, microcysts may be susceptible to certain therapies that could first improve GFR and only later change measurable total kidney volume by slowing microcyst progression to macrocysts either directly or through specialized cells that may be sensitive to rapamycin.
CURRENT MANAGEMENT OF ADPKD
Blood pressure control is essential—but too low is not good. For adult patients with hypertension caused by ADPKD, an acceptable blood pressure range is 120–130/70–80 mm Hg. However, further information from recently published blood pressure guidelines54 and the results of the Halt Progression of Polycystic Kidney Disease (HALT-PKD) study to be reported in late 201455 may provide more precise ranges for blood pressure control in ADPKD.
Although substantial experimental evidence exists for the benefits of inhibiting the up-regulation of the renin-angiotensin-aldosterone system in ADPKD, clinical proof is not yet available to confirm that angiotensin-converting enzyme (ACE) inhibitors or angiotensin receptor blockers (ARBs) are preferred therapy.55 This may be determined by results of the HALT-PKD study, due for release in late 2014.55
Controlling blood pressure should be done with caution. Patients with low GFRs whose blood pressure is too low tend to have a more rapid decline of GFR, as suggested in the Modification of Diet in Renal Disease (MDRD) study in 1995.56
Experimental evidence suggests that avoiding calcium channel blockers may be advisable. Yamaguchi et al34 found that calcium channel blockers worsen the calcium transport defect and convert tubular epithelial cells to a proliferative phenotype.34
High fluid intake (2,500–3,000 mL/day), because it suppresses vasopressin, may be useful if permitted by several factors such as the patient’s cardiopulmonary and renal and electrolyte status, other medications, and diet.31 The reader is referred to a detailed description of the precautions necessary when prescribing high water intake.31 Patients should have their fluid intake managed by a physician and their renal function and serum sodium and electrolytes monitored regularly in order to avoid hyponatremia. Severe hyponatremia has occurred in patients with ADPKD and impaired kidney function who drank excessive quantities of water. Cardiac and pulmonary complications from excessive fluid intake are also possible, especially with a low GFR and compromised cardiac function.
A low-sodium diet, if not a contributing factor in hyponatremia, can be used under physician direction in the management of hypertension as well as in the prevention of calcium oxalate kidney stones.
Caffeine should be avoided because it may interfere with the activity of the phosphodiesterase that is necessary for the catabolism of cAMP to 5′AMP.
A low-protein diet is of unproven benefit,56 but it is prudent to avoid high protein intake.57
Complications such as bleeding (into or from cysts), infection (urinary tract, kidney cysts, and liver cysts), kidney stones, and urinary tract obstruction should be treated promptly and may require hospitalization.
Regular symptom reviews and physical examinations need to be performed with nonrenal concerns also in mind, such as intracranial aneurysms and cardiac valve lesions.11,58
Formal genetic counseling and molecular testing are becoming more frequently indicated as more complex inheritance patterns arise.6–8,59
Renal replacement therapy in the form of dialysis or transplantation is usually available for the patient when end-stage renal disease occurs. In the largest study thus far, ADPKD patient survival with a kidney transplant was similar to that of patients without ADPKD (about 93% at 5 years), and from 5 years to 15 years death-censored graft survival was actually better.60 Thromboembolic events are more frequent after transplantation,27,60 but they may also occur before transplantation from a massive right kidney compressing the iliac vein or the inferior vena cava, or both, leading to thrombus formation.26
Investigational as well as standard drug studies have intensified. Results from a large randomized study in approximately 1,000 adult ADPKD patients that evaluated over 6 to 8 years the effects of ACE inhibition with or without ARB treatment of hypertension, at both usual and lower blood pressure ranges in those with good renal function, are expected in late 2014.55 Outcomes from a few small clinical studies, eg, one with long-acting somatostatin31,61 and one using low-dose rapamycin48 in adults with ADPKD, will require confirmation in large randomized placebo-controlled clinical studies. In a new 3-year randomized placebo-controlled study of 91 children and young adults (ages 8 to 22) with ADPKD and essentially normal renal function who continued treatment with lisinopril, the addition of pravastatin (20 mg or 40 mg daily based on age) resulted in a significant reduction in the number of patients (46% vs 68%, respectively, P = .03) experiencing a greater than 20% change (increase) in height-adjusted total kidney volume.62 Change in GFR was not reported,62 but an earlier 4-week study in 10 patients treated with simvastatin did show an increase in renal blood flow and GFR.63 Numerous other agents that lack human studies include some described in older experimental work (eg, amiloride,31,64 citrate31,65) and many others from a growing list of potential therapeutic targets.31,66–73 It must be emphasized that there is no FDA-approved medication specifically for the treatment of ADPKD.
Future specific treatments of ADPKD may also involve minimally toxic doses of combination or sequential therapy directed at precystic and then both micro- and macrocystic/cystic fluid expansion aspects of ADPKD.48,74 Unfortunately, at the present time there is no specific FDA-approved therapy for ADPKD.
- Torres VE, Harris PC. Mechanisms of disease: autosomal dominant and recessive polycystic kidney diseases. Nat Clin Pract Nephrol 2006; 2:40–55.
- Torres VE, Harris PC. Autosomal dominant polycystic kidney disease: the last 3 years. Kidney Int 2009; 76:149–168.
- United States Renal Data System. 2013 atlas of CKD & ESRD. Volume 2 - atlas ESRD:172. www.usrds.org/atlas.aspx. Accessed June 4, 2014.
- Barua M, Cil O, Paerson AD, et al. Family history of renal disease severity predicts the mutated gene in ADPKD. J Am Soc Nephrol 2009, 20:1833–1838.
- Harris PC, Bae KT, Rossetti S, et al. Cyst number but not the rate of cystic growth is associated with the mutated gene in autosomal dominant polycystic kidney disease. J Am Soc Nephrol 2006; 17:3013–3019.
- Vujic M, Heyer CM, Ars E, et al. Incompletely penetrant PKD1 alleles mimic the renal manifestations of ARPKD. J Am Soc Nephrol 2010; 21:1097–1102.
- Harris PC. What is the role of somatic mutation in autosomal dominant polycystic kidney disease? J Am Soc Nephrol 2010; 21:1073–1076.
- Watnick T, He N, Wang K, et al. Mutations of PKD1 in ADPKD2 cysts suggest a pathogenic effect of trans-heterozygous mutations. Nat Genet 2000; 25:143–144.
- Ravine D, Gibson RN, Walker RG, Sheffield LJ, Kincaid-Smith P, Danks DM. Evaluation of ultrasonographic diagnostic criteria for autosomal dominant polycystic kidney disease 1. Lancet 1994; 343:824–827.
- Pei Y, Obaji J, Dupuis A, et al. Unified criteria for ultrasonographic diagnosis of ADPKD. J Am Soc Nephrol 2009; 20:205–212.
- Torres VE, Harris PC, Pirson Y. Autosomal dominant polycystic kidney disease. Lancet 2007; 369:1287–1301.
- Bajwa ZH, Sial KA, Malik AB, Steinman TI. Pain patterns in patients with polycystic kidney disease. Kidney Int 2004; 66:1561–1569.
- Jouret F, Lhommel R, Beguin C, et al. Positron-emission computed tomography in cyst infection diagnosis in patients with autosomal dominant polycystic kidney disease. Clin J Am Soc Nephrol 2011; 6:1644–1650.
- Nishiura JL, Neves RF, Eloi SR, Cintra SM, Ajzen SA, Heilberg IP. Evaluation of nephrolithiasis in autosomal dominant polycystic kidney disease patients. Clin J Am Soc Nephrol 2009; 4:838–844.
- Hiura T, Yamazaki H, Saeki T, et al. Nephrotic syndrome and IgA nephropathy in polycystic kidney disease. Clin Exp Nephrol 2006; 10:136–139.
- Hossack KF, Leddy CL, Johnson AM, Schrier RW, Gabow PA. Echocardiographic findings in autosomal dominant polycystic kidney disease. N Engl J Med 1988; 319:907–912.
- Rossetti S, Chauveau D, Kubly V, et al. Association of mutation position in polycystic kidney disease 1 (PKD1) gene and development of a vascular phenotype. Lancet 2003; 361:2196–2201.
- Linn FH, Wijdicks EF, van der Graaf Y, Weerdesteyn-van Vliet FA, Bartelds AI, van Gijn J. Prospective study of sentinel headache in aneurismal subarachnoid haemorrhage. Lancet 1994; 344:590–593.
- Belz MM, Fick-Brosnahan GM, Hughes RL, et al. Recurrence of intracranial aneurysms in autosomal-dominant polycystic kidney disease. Kidney Int 2003; 63:1824–1830.
- Irazabal MV, Huston J, Kubly V, et al. Extended follow-up of unruptured intracranial aneurysms detected by presymptomatic screening in patients with autosomal dominant polycystic kidney disease. Clin J Am Soc Nephrol 2011; 6:1274–1285.
- Salman A-S, White PM, Counsell CE, et al; Scottish Audit of Intracranial Vascular Malformations Collaborators. Outcome after conservative management or intervention for unruptured brain arteriovenous malformations. JAMA 2014; 311:1661–1669.
- Vijay A, Vijay A, Pankaj P. Autosomal dominant polycystic kidney disease: a comprehensive review. Nephrourol Mon 2010; 2:172–192.
- Grantham JJ, Torres VE, Chapman AB, et al; CRISP Investigators. Volume progression in polycystic kidney disease. N Engl J Med 2006; 354:2122–2130.
- Bae KT, Grantham JJ. Imaging for the prognosis of autosomal dominant polycystic kidney disease. Nat Rev Nephrol 2010; 6:96–106.
- van den Dool SW, Wasser NM, de Fijter JW, Hoekstra J, van der Geest RJ. Functional renal volume: quantitative analysis at gadolinium-enhanced MR angiography—feasibility study in healthy potential kidney donors. Radiology 2005; 236:189–195.
- O’Sullivan DA, Torres VE, Heit JA, Liggett S, King BF. Compression of the inferior vena cava by right renal cysts: an unusual cause of IVC and/or iliofemoral thrombosis with pulmonary embolism in autosomal dominant polycystic kidney disease. Clin Nephrol 1998; 49:332–334.
- Tveit DP, Hypolite I, Bucci J, et al. Risk factors for hospitalizations resulting from pulmonary embolism after renal transplantation in the United States. J Nephrol 2001; 14:361–368.
- Pei Y. A “two-hit” model of cystogenesis in autosomal dominant polycystic kidney disease? Trends Mol Med 2001; 7:151–156.
- Qian F, Germino GG. “Mistakes happen”: somatic mutation and disease. Am J Hum Genet 1997; 61:1000–1005.
- Takakura A, Contrino L, Zhou X, et al. Renal injury is a third hit promoting rapid development of adult polycystic kidney disease. Hum Mol Genet 2009; 18:2523–2531.
- Torres VE, Harris PC. Strategies targeting cAMP signaling in the treatment of polycystic kidney disease. J Am Soc Nephrol 2014; 25:18–32.
- Nauli SM, Alenghat FJ, Luo Y, et al. Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat Genet 2003; 33:129–137.
- Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N Engl J Med 2011; 364:1533–1543.
- Yamaguchi T, Wallace DP, Magenheimer BS, Hempson SJ, Grantham JJ, Calvet JP. Calcium restriction allows cAMP activation of the B-Raf/ERK pathway, switching cells to a cAMP-dependent growth-stimulated phenotype. J Biol Chem 2004; 279:40419–40430.
- Verghese E, Ricardo SD, Weidenfeld R, et al. Renal primary cilia lengthen after acute tubular necrosis. J Am Soc Nephrol 2009; 20:2147–2153.
- Wang X, Wu Y, Ward CJ, Harris PC, Torres VE. Vasopressin directly regulates cyst growth in polycystic kidney disease. J Am Soc Nephrol 2008; 19:102–108.
- Torres VE. Cyclic AMP, at the hub of the cystic cycle. Kidney Int 2004; 66:1283–1285.
- Nagao S, Nishii K, Katsuyama M, et al. Increased water intake decreases progression of polycystic kidney disease in the PCK rat. J Am Soc Nephrol 2006; 17:2220–2227.
- Grantham JJ. Therapy for polycystic kidney disease? It’s water, stupid! J Am Soc Nephrol 2008; 19:1–7.
- Wang CJ, Creed C, Winklhofer FT, Grantham JJ. Water prescription in autosomal dominant polycystic kidney disease: a pilot study. Clin J Am Soc Nephrol 2011; 6:192–197.
- Gattone VH, Wang X, Harris PC, Torres VE. Inhibition of renal cystic disease development and progression by a vasopressin V2 receptor antagonist. Nat Med 2003; 9:1323–1326.
- Torres VE, Chapman AB, Devuyst O, et al; TEMPO 3:4 Trial Investigators. Tolvaptan in patients with autosomal dominant polycystic kidney disease. N Engl J Med 2012; 367:2407–2418.
- Shillingford JM, Murcia NS, Larson CH, et al. The mTOR pathway is regulated by polycystin-1, and its inhibition reverses renal cystogenesis in polycystic kidney disease. Proc Natl Acad Sci U S A 2006; 103:5466–5471.
- Hartman TR, Liu D, Zilfou JT, et al. The tuberous sclerosis proteins regulate formation of the primary cilium via a rapamycin-insensitive and polycystin 1-independent pathway. Hum Mol Genet 2009; 18:161–163.
- Serra AL, Poster D, Kistler AD, et al. Sirolimus and kidney growth in autosomal dominant polycystic kidney disease. N Engl J Med 2010; 363:820–829.
- Walz G, Budde K, Mannaa M, et al. Everolimus in patients with autosomal dominant polycystic kidney disease. N Engl J Med 2010; 363:830–840. Errata in: N Engl J Med 2010; 363:1190 and N Engl J Med 2010; 363:1977.
- Walz G, Budde K, Eckardt K-U. mTOR inhibitors and autosomal dominant polycystic kidney disease (correspondence). N Engl J Med 2011; 364:287–288.
- Braun WE, Schold JD, Stephany BR, Spinko RA, Herfs BR. Low dose rapamycin (sirolimus) effects in autosomal dominant polycystic kidney disease: an open-label randomized control pilot study. Clin J Am Soc Nephrol 2014; 9:881–888.
- Karihaloo A, Koraishy F, Huen SC, et al. Macrophages promote cyst growth in polycystic kidney disease. J Am Soc Nephrol 2011; 22:1809–1814.
- Fox R, Nhan TQ, Law GL, Morris DR, Liles WC, Schwartz SM. PSGL-1 and mTOR regulate translation of ROCK-1 and physiological functions of macrophages. EMBO J 2007; 26:505–515. Erratum in: EMBO J 2007; 26:2605.
- Carvalhosa R, Deambrosis I, Carrera P, et al. Cystogenic potential of CD133+ progenitor cells of human polycystic kidneys. J Pathol 2011; 225:129–141.
- Grantham JJ, Mulamalla S, Grantham CJ, et al. Detected renal cysts are tips of the iceberg in adults with ADPKD. Clin J Am Soc Nephrol 2012; 7:1087–1093.
- Grantham JJ, Cook LT, Wetzel LH, Cadnapaphornchai MA, Bae KT. Evidence of extraordinary growth in the progressive enlargement of renal cysts. Clin J Am Soc Nephrol 2010; 5:889–896.
- James PA, Oparil S, Carter BL, et al. 2014 evidence-based guideline for the management of high blood pressure in adults: report from the panel members appointed to the Eighth Joint National Committee (JNC 8). JAMA 2014; 311:507–520.
- Chapman AB, Torres VE, Perrone RD, et al. The HALT polycystic kidney disease trials: design and implementation. Clin J Am Soc Nephrol 2010; 5:102–109.
- Klahr S, Breyer JA, Beck GJ, et al. Dietary protein restriction, blood pressure control, and the progression of polycystic kidney disease. Modification of Diet in Renal Disease Study Group. J Am Soc Nephrol 1995; 5:2037–2047.
- Thilly N. Low-protein diet in chronic kidney disease: from questions of effectiveness to those of feasibility. Nephrol Dial Transplant 2013; 28:2203–2205.
- Luciano RL, Dahl NK. Extra-renal manifestations of autosomal dominant polycystic kidney disease (ADPKD): considerations for routine screening and management. Nephrol Dial Transplant 2014; 29:247–254.
- Harris PC, Rossetti S. Molecular diagnostics for autosomal dominant polycystic kidney disease. Nat Rev Nephrol 2010; 6:197–206.
- Jacquet A, Pallet N, Kessler M, et al. Outcomes of renal transplantation in patients with autosomal dominant polycystic kidney disease: a nationwide longitudinal study. Transpl Int 2011; 24:582–587.
- Ruggenenti P, Remuzzi A, Ondei P, et al. Safety and efficacy of long-acting somatostatin treatment in autosomal-dominant polycystic kidney disease. Kidney Int 2005; 68:206–216.
- Cadnapaphornchai MA, George DM, McFann K, et al. Effect of pravastatin on total kidney volume, left ventricular mass index, and microalbuminuria in pediatric autosomal dominant polycystic kidney disease. Clin J Am Soc Nephrol 2014; 9:889–896.
- van Dijk MA, Kamper AM, van Veen S, Souverjin JH, Blauw GJ. Effect of simvastatin on renal function in autosomal dominant polycystic kidney disease. Nephrol Dial Transplant 2001; 16:2152–2157.
- Grantham JJ, Uchich M, Cragoe EL, et al. Chemical modification of cell proliferation and fluid secretion in renal cysts. Kidney Int 1989; 35:1379–1389.
- Tanner GA. Potassium citrate/citric acid intake improves renal function in rats with polycystic kidney disease. J Am Soc Nephrol 1998; 9:1242–1248.
- Belibi FA, Edelstein CL. Novel targets for the treatment of autosomal dominant polycystic kidney disease. Expert Opin Investig Drugs 2010; 19:315–328.
- Tao Y, Kim J, Yin Y, et al. VEGF receptor inhibition slows the progression of polycystic kidney disease. Kidney Int 2007; 72:1358–1366.
- Terryn S, Ho A, Beauwens R, Devuyst O. Fluid transport and cystogenesis in autosomal dominant polycystic kidney disease. Biochim Biophys Acta 2011; 1812:1314–1321.
- Thiagarajah JR, Verkman AS. CFTR inhibitors for treating diarrheal disease. Clin Pharmacol Ther 2012; 92:287–290.
- Boehn SN, Spahn S, Neudecker S, et al. Inhibition of Comt with tolcapone slows proression of polycystic kidney disease in the more severely affected PKD/Mhm (cy/+) substrain of the Hannover Sprague-Dawley rat. Nephrol Dial Transplant 2013; 28:2045–2058.
- Rees S, Kittikulsuth W, Roos K, Strait KA, Van Hoek A, Kohan DE. Adenylyl cyclase 6 deficiency ameliorates polycystic kidney disease. J Am Soc Nephrol 2014; 25:232–237.
- Buchholz B, Schley G, Faria D, et al. Hypoxia-inducible factor-1a causes renal cyst expansion through calcium-activated chloride secretion. J Am Soc Nephrol 2014; 25:465–474.
- Wallace DP, White C, Savinkova L, et al. Periostin promotes renal cyst growth and interstitial fibrosis in polycystic kidney disease. Kidney Int 2014; 85:845–854.
- Leuenroth SJ, Crews CM. Targeting cyst initiation in ADPKD. J Am Soc Nephrol 2009; 20:1–3.
Autosomal dominant polycystic kidney disease (ADPKD) is the most common inherited renal disease, has an estimated prevalence of 1:400 to 1:1,000 live births in the United States, and occurs worldwide.1,2 There are about 700,000 people living with it in the United States, and about 6,000 new cases arise annually. It accounts for nearly 5% of all patients with end-stage renal disease in the United States.3
This paper will offer an overview of the pathogenesis of renal cysts, review some of the clinical aspects of ADPKD including diagnosis and management of complications, and discuss recent drug trials and current management.
TWO TYPES—PKD1 IS MORE COMMON AND PROGRESSES MORE RAPIDLY
Two major forms of ADPKD are recognized and can usually be determined by genetic testing: PKD1, accounting for about 85% of cases, and PKD2, accounting for 15%.
The gene locus for PKD1 is on the short arm of the 16th chromosome (16p13.3), and its glycoprotein gene product is polycystin 1 (PC1), a large molecule with 4,303 amino acids.2 PC1 has a long N-terminal extracellular tail that can function as a mechanosensor. Disease progression is much faster with PKD1, and end-stage renal disease usually occurs before age 56.4
In PKD2, the gene locus is on the long arm of the fourth chromosome (4q21–23), and has a smaller glycoprotein gene product, polycystin 2 (PC2), that plays a role in calcium transport. The disease course of PKD2 tends to be slower. End-stage renal disease might not develop in the patient’s lifetime, since it typically develops when the patient is more than 70 years old.4
Although the growth rate of renal cysts is similar between the two types, patients with PKD1 develop about twice as many cysts as those with PDK2, and their cyst development starts at a younger age.5
Typically, patients have a clear phenotype and a positive family history, but in about 10% of possible ADPKD cases, there is no family history of ADPKD. Genetic variations such as incompletely penetrant PKD1 alleles,6 hypomorphic alleles,7 and trans-heterozygous mutations8 account for at least some of these cases.
IMAGING CRITERIA HAVE BROADENED
Ultrasonographic criteria for the diagnosis of ADPKD that were published in 1994 were based on patients who had a family history of PKD1.9 The criteria have since been modified (the “unified criteria”) to include patients with a family history of PKD2 who begin cyst development at a later age and with lower numbers.10 For patients ages 30 to 39, a previously difficult diagnostic group, the criterion for the minimum number of cysts visible on ultrasonography changed from four to three, improving the sensitivity of detecting disease from approximately 76% to approximately 95% (Table 1).9,10 It is important to note that these criteria apply only to patients “at risk,” ie, with a positive family history of ADPKD.
Computed tomography (CT) and magnetic resonance imaging (MRI) classically show bilaterally enlarged multicystic kidneys, though variations can be seen.
DISEASE CAN PRESENT IN MYRIAD WAYS
Although cystic kidney disease is the basic underlying problem, undiagnosed patients may present with a variety of symptoms caused by other manifestations of ADPKD (Table 2).
Hypertension is the most common presentation, occurring in about 50% of patients ages 20 to 34, and essentially 100% of those with end-stage renal disease.11 It is associated with up-regulation of the renin-angiotensin-aldosterone system.
Pain is typically located in the abdomen, flank, or back and can occur in a localized or diffuse manner. Early abdominal distress is often simply described as “fullness.” Localized pain is usually caused by bleeding into or rupture of a cyst, renal stones, or infection.12 Because renal cysts are noncommunicating, bleeding can occur into a cyst and cause pain without gross hematuria. Compression by greatly enlarged kidneys, liver, or both can cause a variety of gastrointestinal symptoms such as reflux esophagitis and varying degrees of constipation. Diffuse pain is often musculoskeletal and related to exaggerated lordosis from increasing abdominal size due to enlarging cystic kidneys and sometimes liver.12 In carefully selected cases, cyst aspiration may be helpful.11
Although renal carcinomas are rare and not more frequent than in the general population, they can occur at an earlier age and with constitutional symptoms.11
Urinary tract infections are increased in frequency. A patient may have a simple urinary tract infection that is cured with the appropriate antibiotic. However, a urinary tract infection repeatedly recurring with the same organism is a strong clue that an infected cyst is the source and requires more intensive treatment with the appropriate cyst-penetrating antibiotic. On the other hand, because cysts are noncommunicating, an infected cyst might be present despite a negative urine culture.
Identifying infected cysts can be a challenge with conventional imaging techniques, but combined positron emission tomography and CT (PET/CT) can be a valuable though expensive diagnostic tool to identify an infected kidney or liver cyst, or to identify an unsuspected source of the pain and infection.13
Jouret et al13 evaluated 27 PET/CT scans performed in 24 patients with ADPKD and suspicion of an abdominal infection. Patients were deemed to have probable cyst infection if they met all of the following criteria: temperature more than 38°C for longer than 3 days, loin or liver tenderness, plasma C-reactive protein level greater than 5 mg/dL, and no evidence of intracystic bleeding on CT. Patients with only two or three of these criteria were classified as having fever of unknown origin. Diagnosis of cyst infection was confirmed by cyst fluid analysis.
PET/CT identified a kidney or liver cyst infection in 85% of 13 infectious events in 11 patients who met all the criteria for probable cyst infection; CT alone contributed to the diagnosis in only one patient.13 In those with fever of unknown origin, PET/CT identified a source of infection in 64% of 14 events in 13 patients: two infected renal cysts, as well as one patient each with other infections that would be difficult to diagnose clinically, ie, small bowel diverticulitis, psoas abscess, diverticulitis of the right colon, pyelonephritis in a transplanted kidney, infected abdominal aortic aneurysm, prostatitis, colitis, and Helicobacter pylori gastritis. Results of PET/CT were negative in five patients with intracystic bleeding.
Kidney stones occur in 20% to 36% of patients.11,14 Uric acid stones occur at almost the same frequency as calcium oxalate stones.
Chronic kidney disease not previously diagnosed may be the presenting condition in a small percentage of patients, sometimes those in whom much earlier hypertension was not fully evaluated. ADPKD is typically not associated with significant proteinuria (eg, nephrotic range), and the presence of heavy proteinuria usually indicates the presence of a superimposed primary glomerulopathy.15
Cysts in other locations. By MRI, liver cysts are present in 58% of patients ages 15 to 24, rising to 94% in those ages 35 to 46.11 Because liver cysts are estrogen-dependent, they are more prominent in women. A small percentage of patients develop cysts in the pancreas (5%), arachnoid membranes (8%), and seminal vesicles (40% of men with ADPKD).11
Cardiovascular abnormalities occur in almost one-third of patients with ADPKD, usually as mitral and aortic valve abnormalities.16 Aneurysms of the aortic root and abdominal aorta can also occur, in addition to intracranial aneurysms (see below).17
Intracranial aneurysms are not uncommon, and size usually determines their risk.
Intracranial aneurysms are strongly influenced by family history: 16% of ADPKD patients with a family history of intracranial aneurysm also develop them, compared with 5% to 6% of patients with no family history.11 The anterior cerebral circulation is involved in about 80% of cases. A sentinel or sudden “thunderclap” headache is a classic presentation that may precede full-blown rupture in about 17% of cases.18 Patients who rupture an intracranial aneurysm have a mean age of 39, usually have normal renal function, and can be normotensive.11
For patients with no history of subarachnoid hemorrhage, the 5-year cumulative rupture rates for patients with aneurysms located in the internal carotid artery, anterior communicating or anterior cerebral artery, or middle cerebral artery were 0% for aneurysms less than 7 mm, 2.6% for those 7 to 12 mm, 14.5% for those 13 to 24 mm, and 40% for those 25 mm or larger, with higher rates for the same sizes in the posterior circulation.11
In patients without symptoms, size is correlated with risk of rupture: less than 4 mm is usually associated with very low risk, 4 to less than 7 mm with moderate risk, and 7 mm or more with increasing risk. An aneurysm larger than 10 mm is associated with roughly a 1% risk of rupture per year.19
Irazabal et al20 retrospectively studied 407 patients with ADPKD who were screened for intracranial aneurysm. Saccular aneurysms were detected in 45 patients; most were small (median diameter 3.5 mm). During cumulative imaging follow-up of 243 years, only one new intracranial aneurysm was detected (increasing from 2 to 4.4 mm over 144 months) and two previously identified aneurysms grew (one increasing 4.5 to 5.9 mm over 69 months and the other 4.7 to 6.2 mm over 184 months). No change occurred in 28 patients. Seven patients were lost to follow-up, however. During cumulative clinical follow-up of 316 years, no aneurysm ruptured. Two patients were lost to follow-up, three had surgical clipping, and five died of unrelated causes. The authors concluded that presymptomatic intracranial aneurysms are usually small, and that growth and rupture risks are no higher than for unruptured intracranial aneurysms in the general population. A 2014 study also suggests a conservative approach for managing intracranial aneurysm in the general population.21
In asymptomatic ADPKD patients, it is reasonable to reserve screening for those with a positive family history of intracranial aneurysm or subarachnoid hemorrhage, those with a previous ruptured aneurysm, those in high-risk professions (eg, pilots), and for patients prior to anticoagulant therapy or major surgery possibly associated with hemodynamic instability.11,22 Certain extremely anxious patients might also need to be studied. Screening can be performed with magnetic resonance angiography without gadolinium contrast. It is prudent to have patients with an intracranial aneurysm thoroughly evaluated by an experienced neurosurgeon with continued follow-up.
PROGRESSION OF ADPKD
The Consortium for Radiologic Imaging Studies of Polycystic Kidney Disease (CRISP) study23 evaluated 241 patients with ADPKD (ages 15 to 46) by measuring the annual rate of change in total kidney volume, total cyst volume, and iothalamate glomerular filtration rate (GFR) over 3 years. The annual increase in total kidney volume averaged 5.3%,23 though the reported range with various imaging techniques is from 4% to 12.8% in adults.24 This study focused on macrocystic disease, ie, cysts that are visible by MRI and measurably increase total kidney volume. Although larger total kidney volume at baseline generally predicted a more rapid decline in GFR, there were wide and overlapping variations in yearly GFR declines within and among different total-kidney-volume groups.23
SPECIAL CLINICAL PROBLEMS IN ADPKD
Case 1: A man with ADPKD develops new and increasing proteinuria
A 55-year-old man with ADPKD and stage 3 chronic kidney disease developed new and increasing proteinuria, rising to 5,500 mg per 24 hours. What is the most likely explanation?
- Rapidly progressive renal failure with increasing proteinuria in ADPKD
- Bilateral renal vein thromboses because of cyst compression
- Malignant hypertension with bilateral renal artery compression
- Superimposed primary glomerulopathy
- Multiple infected renal cysts with pyonephrosis
Answer: Superimposed primary glomerulopathy.
ADPKD (similar to uncomplicated obstructive uropathy, pyelonephritis, main renal artery disease, and often cases of interstitial nephritis without secondary glomerular changes) typically does not result in nephrotic-range proteinuria. A superimposed primary glomerulopathy, focal segmental glomerulosclerosis, was the biopsy-proved diagnosis.
At least 21 cases have been reported of AD-PKD with nephrotic-range proteinuria and a renal biopsy showing a primary glomerulopathy, including focal segmental glomerulosclerosis (5 cases), minimal-change disease (5), membranous nephropathy (3), IgA nephropathy (2), and one each of crescentic glomerulonephropathy, diabetic nephropathy, membranoproliferative glomerulonephritis, postinfectious glomerulonephropathy, amyloid glomerulopathy, and mesangioproliferative glomerulopathy.15 Treatment was directed at the primary glomerulopathy, and the outcomes corresponded to the primary diagnosis (eg, with appropriate treatment, three of the five patients with focal segmental glomerulosclerosis progressed to end-stage renal disease, all of the patients with minimal-change disease went into remission, and one of the two cases with IgA nephropathy improved).15
Case 2: A woman with ADPKD and advanced renal failure develops shortness of breath
A 47-year-old woman with very large polycystic kidneys (total kidney volume 7,500 mL; normal range for a single kidney approximately 136–295 mL, mean 196)25 and estimated GFR of 25 mL/min developed new-onset shortness of breath while climbing steps and later even when making a bed. She had no chest pain, cough, or edema. She was sent directly to the emergency department and was admitted and treated; her condition improved, and she was discharged after 6 days. What did she have?
- Presentation of rare cystic pulmonary disease in ADPKD
- Onset of pneumonia with early bacteremia
- Progressive reduction in ventilatory capacity from massive polycystic kidneys and liver elevating both sides of the diaphragm
- Pulmonary emboli from an iliac vein or inferior vena cava source
- Progressive anemia accompanying rapidly worsening stage 4 chronic kidney disease
Answer: She had pulmonary emboli from an iliac vein (right) or inferior vena cava source.
Pulmonary emboli in ADPKD can be caused by thrombi in the inferior vena cava or the iliac or femoral vein because of compression by a massive right polycystic kidney. Four cases were reported at Mayo Clinic,26 three diagnosed by MRI and one with CT. One additional case occurred at Cleveland Clinic. All patients survived after treatment with anticoagulation therapy; early nephrectomy was required in two cases.
Interestingly, following kidney transplantation, the patients at greatest risk for pulmonary emboli are those with ADPKD as their original disease.27
RENAL CYSTS RESULT FROM COMBINED MUTATIONS, INJURY
The germline ADPKD mutation that occurs in one allele of all renal tubular epithelial cells is necessary but not sufficient for cystogenesis.28 One or more additional somatic mutations of the normal allele—the “second hit”—also develop within individual tubular epithelial cells.28,29 These epithelial cells undergo clonal proliferation, resulting in tubular dilatation and cyst formation. Monoclonality of cells in cysts has been documented.
Ischemia-reperfusion injury can be viewed as a “third hit.”30 In PKD1 knockout mice, which at 5 weeks of age normally develop only mild cystic kidney disease, the superimposition of unilateral ischemia-reperfusion injury at 8 weeks caused widespread and rapid cyst formation. It is believed that acute renal injury reactivates developmental signaling pathways within 48 hours that trigger epithelial cell proliferation and then cyst development detectable by MRI 2 weeks later. Although this phenomenon has not been documented in humans, it is a cautionary tale.
CYSTOGENESIS INVOLVES MULTIPLE PATHWAYS
A comprehensive description of pathways leading to renal cyst formation is beyond the scope of this article, and the reader is referred to much more detailed and extensive reviews.2,31 Disturbances in at least three major interconnected pathways promote cystogenesis in renal tubular epithelial cells:
- Normal calcium transport into the endoplasmic reticulum is disrupted by abnormal polycystins on the surface of the primary cilium
- Vasopressin and other stimuli increase the production of cyclic adenosine monophosphate (cAMP)
- The mammalian target of rapamycin (mTOR) proliferative pathway is up-regulated.
DISRUPTION OF CALCIUM TRANSPORT IN THE PRIMARY CILIUM
Primary cilia are nonmotile cellular organelles of varying size, from about 0.25 μm up to about 1 μm.32 Each primary cilium has nine peripheral pairs of microtubules but lacks a centrally located pair that is present in motile cilia. Primary cilia are ubiquitous and have been highly conserved throughout evolution. A single cilium is present on almost all vertebral cells.33
Cilial defects have been identified in autosomal dominant as well as recessive diseases and are known as ciliopathies.33 Although rare in humans, they can affect a broad spectrum of organs other than the kidney, including the eye, liver, and brain.33
Urine flow in a renal tubule is believed to exert mechanical stimulation on the extracellular flagellum-like N-terminal tail of PC1 that extends from a primary cilium into the urinary space. PC1 in concert with PC2 opens PC2 calcium channels, allowing calcium ions to flow down the microtubules to ryanodine receptors and the basal body.32,33 This leads to local release of calcium ions that regulate cell proliferation.32,34 However, in ADPKD kidneys, PC1 and PC2 molecules are sparse or mutated, resulting in defective calcium transport, increased and unregulated tubular epithelial cell proliferation, and cyst formation.
In a totally different clinical setting, biopsies of human renal transplants that sustained acute tubular necrosis during transplantation reveal that a cilium dramatically elongates in response to injury,35 possibly as a compensatory mechanism to maintain calcium transport in the presence of meager urine flow and to restore the proliferation of tubular epithelial cells in a regulated repair process.
THE ROLE OF VASOPRESSIN AND ACTIVATION OF cAMP
In classic experiments, Wang et al36 cross-bred rats having genetically inherited polycystic kidney disease (actually, autosomal recessive polycystic kidney disease) with Brattleboro rats that completely lack vasopressin. At 10 and 20 weeks of age, the offspring had virtually complete inhibition of cystogenesis because of the absence of vasopressin. However, when vasopressin was restored by exogenous administration continuously for 8 weeks, the animals formed massive renal cysts.
Vasopressin activates cAMP, which then functions as a second messenger in cell signaling. cAMP increases the activation of the protein kinase A (PKA) pathway, which in turn increases downstream activity of the B-raf/ERK pathway. Up-regulation of cAMP and PKA appears to perpetuate activation of canonical Wnt signaling, down-regulate non-canonical Wnt/planar cell polarity signaling, and lead to loss of tubular diameter control, resulting in cyst formation.31 Normally, cAMP is degraded by phosphodiesterase. However, because of the primary cilium calcium transport defect in ADPKD, phosphodiesterase is reduced and cAMP persists.37 In conjunction with the defective primary cilial calcium transport, cAMP exerts a proliferative effect on renal tubular epithelial cells that is opposite to its effect in normal kidneys.31,32 cAMP also up-regulates the cystic fibrosis transmembrane conductance regulator (CFTR) that promotes chloride ion transport. Sodium ions follow the chloride ions, leading to fluid accumulation and cyst enlargement.31
Inhibiting vasopressin by increasing water intake
A simple key mechanism for limiting vasopressin secretion is by sufficient water ingestion. Nagao et al38 found that rats with polycystic kidney disease given water with 5% glucose (resulting in 3.5-fold increased fluid intake compared with rats given tap water) had a 68% reduction in urinary vasopressin and a urine osmolality less than 290 mOsm/kg. The high-water-intake rats had dramatically reduced cystic areas in the kidney and a 28% reduction of kidney-to-body weight ratio vs controls.
In an obvious oversimplification, these findings raised the question of whether a sufficient increase in water intake could be an effective therapy for polycystic kidney disease.39 A pilot clinical study evaluated changes in urine osmolality in eight patients with ADPKD who had normal renal function.40 At baseline, 24-hour urine osmolality was typically elevated to approximately 753 mOsm/kg compared to the plasma at 285 mOsm/kg, indicating that antidiuresis is the usual state. During the 2-week study, urine volume and osmolality were measured, and additional water intake was adjusted in order to achieve a urine osmolality goal of 285 ± 45 mOsm/kg. These adjustments resulted in water intake that appeared to be in the range of 2,400 to 3,000 mL per 24 hours. The major limitations of the study were that it was very short term, and there was no opportunity to measure changes in total kidney volume or estimated GFR.
In a recent preliminary report from Japan, high water intake (2,500–3,000 mL daily) in 18 ADPKD patients was compared over 12 months with ad libitum water intake in 14 ADPKD controls (clinicaltrials.gov NCT 01348505). There was no statistically significant change in total kidney volume or cystatin-estimated GFR in those on high water intake, but serious defects in study design (patients in the high water intake group were allowed to decrease their intake if it was causing them difficulty, and patients in the ad libitum water intake group had no measurement of their actual water intake) prevent any conclusions because there was no evidence that the groups were different from one another with respect to the key element of the study, namely, water intake.
Blocking the vasopressin receptor slows disease progression
Using another approach, Gattone et al41 inhibited the effect of vasopressin by blocking the vasopressin 2 receptor (V2R) in mouse and rat models of polycystic kidney disease, using an experimental drug, OPC31260. The drug halted disease progression and, in one situation, appeared to cause regression of established disease. As noted by Torres and Harris,31 even though both increased water intake and V2R antagonists decrease cAMP in the distal tubules and collecting ducts, circulating levels of vasopressin are decreased by increased water intake but increased by V2R antagonists.
After these remarkable results in animal models, clinical trials of the V2R antagonist tolvaptan were conducted in patients with ADPKD. In the Tolvaptan Efficacy and Safety in Management of Autosomal Dominant Polycystic Kidney Disease and Its Outcomes 3:4 study,42 1,445 adults (ages 18 to 50) with ADPKD in 133 centers worldwide were randomized to receive either tolvaptan or placebo for 3 years. Key inclusion criteria included good renal function (estimated GFR ≥ 60 mL/min) and total kidney volume of at least 750 mL (mean 1,700 mL) as measured by MRI. Tolvaptan was titrated to the highest tolerated twice-daily dose (average total of 95 mg/day). All patients were advised to maintain good hydration and to avoid thirst by drinking a glass of water after each urination. Unfortunately, neither water intake nor urine output was measured.
The primary end point was the annual rate of change in total kidney volume, with secondary end points of clinical progression (worsening kidney function, pain, hypertension, albuminuria), and rate of decline in kidney function as measured by the slope of the reciprocal of serum creatinine.42
Patients in the tolvaptan arm had a slower annual increase in total kidney volume than controls (2.8% vs 5.5%, respectively, P < .001) and a slower annual decline in renal function (−2.61 vs −3.81 mg/mL−1, respectively, P < .001).42 More participants in the treatment group withdrew than in the placebo group (23% vs 14%, respectively).
Adverse events occurred more frequently with tolvaptan.42 Liver enzyme elevations of greater than three times the upper limit of normal occurred in 4.4% of patients in the treatment group, leading to a drug warning issued in January 2013. As expected, side effects related to diuresis (urinary frequency, nocturia, polyuria, and thirst) were more frequent in the treatment group, occurring in up to 55% of participants.
The authors noted, “Although maintaining hydration helped ensure that the blinding in the study was maintained, the suppression of vasopressin release in the placebo group may have led to an underestimation of the beneficial effect of tolvaptan and may account for the lower rates of kidney growth observed in the placebo group.”42
In 2013, the US Food and Drug Administration (FDA) denied a new drug application for tolvaptan as a treatment for ADPKD.
THE mTOR PATHWAY IS UP-REGULATED
The mTOR pathway that plays a major role in cell growth and proliferation includes interaction of the cytoplasmic tail of polycystin 1 with tuberin.43 Activation products of mTOR, including phospho-S6K, have been found in tubular epithelial cells lining cysts of ADPKD kidneys but not in normal kidneys.43 Mutant mice with polycystic disease had phospho-S6K in tubular epithelial cells of cysts, whereas those treated with the mTOR inhibitor rapamycin did not.43 But subsequent studies have shown that only a low level of mTOR activation is present in 65% to 70% of ADPKD cysts.44
Two major studies of the treatment of ADPKD with rapamycin that were published contemporaneously in 2010 failed to demonstrate any significant benefit with mTOR inhibitor treatment.45,46
Serra et al45 conducted an 18-month, open-label trial of 100 ADPKD patients ages 18 to 40 with an estimated GFR (eGFR) of at least 70 mL/min. Patients were randomized to receive rapamycin, given as sirolimus 2 mg per day, or standard care. The primary end point was the reduction in the growth rate of total kidney volume, measured by MRI. Secondary end points were eGFR and protein excretion (albumin-creatinine ratio). No significant difference was found in total kidney volume, but a nonsignificant stabilization of eGFR was noted.
Walz et al46 in a 2-year, multicenter, double-blind trial, randomized 433 patients (mean age 44; mean eGFR 54.5 mL/min) to treatment with either the short-acting mTOR inhibitor everolimus (2.5 mg twice daily) or placebo. Although patients in the treatment group had less of an increase in total kidney volume (significant at 1 year but not at 2 years), they tended to show a decline in eGFR. But further analysis showed that the only patients who had a reduction in eGFR were males who already had impaired kidney function at baseline.47
In a pilot study, 30 patients with ADPKD (mean age 49) were randomized to one of three therapies:
- Low-dose rapamycin (trough blood level 2–5 ng/mL)
- Standard-dose rapamycin (trough blood level > 5–8 ng/mL)
- Standard care without rapamycin.48
In contrast to other studies, the primary end point was the change in iothalamate GFR at 12 months, with change in total kidney volume being a secondary end point.
At 12 months, with 26 patients completing the study, the low-dose rapamycin group (n = 9) had a significant increase in iothalamate GFR of 7.7 ± 12.5 mL/min/1.73 m2, whereas the standard-dose rapamycin group (n = 8) had a nonsignificant increase of 1.6 ± 12.1 mL/min/1.73 m2, and the no-rapamycin group (n = 9) had a fall in iothalamate GFR of 11.2 ± 9.1 mL/min/1.73 m2 (P = .005 for low-dose vs no rapamycin; P = .07 for standard-dose vs no rapamycin; P = .52 for low-dose vs standard-dose rapamycin; and P = .002 for combined low-dose and standard-dose rapamycin vs no rapamycin.).48 These differences were observed despite there being no significant change in total kidney volume in any of the groups. Patients on low-dose rapamycin had fewer adverse effects than those on standard dose and were more often able to continue therapy for the entire study. This, and the use of iothalamate GFR rather than eGFR to measure GFR, are believed to be the main reasons that low-dose effects were more pronounced than those with standard doses. One may speculate that rapamycin may have its effect on microcysts and cystogenic cells, resulting in stabilization of or improvement in renal function without detectable slowing in total kidney volume enlargement. Mechanisms for this possibility involve new concepts, as discussed below.
NEW CONCEPTS
Specialized cells also promote renal cyst formation
Specialized cells that promote cyst formation have been identified by Karihaloo et al49 in a mouse model of polycystic kidney disease. In this model, alternatively activated macrophages homed to cystic areas and promoted cyst growth. These findings suggested that interrupting the homing and proliferative signals of macrophages could be a therapeutic target for ADPKD. Although rapamycin can suppress macrophage proliferation by macrophage colony-stimulating factor and inhibit macrophage function,50 alternatively activated macrophages have not been specifically studied for rapamycin responsiveness.
More promising is evidence that CD133+ progenitor cells from human ADPKD kidneys—but not from normal human kidneys—form cysts in vitro and in severe combined immunodeficient mouse models.51 Treatment with rapamycin decreased proliferation of the de-differentiated CD133+ cells from ADPKD patients and reduced cystogenesis.51
Visible cysts are the tip of the iceberg
Using ADPKD nephrectomy specimens from eight patients, Grantham et al52 compared cyst counts by MRI and by histology and found that for every renal cyst detected by MRI, about 62 smaller cysts (< 0.9 mm) are present in the kidney. For a typical patient having an average of 587 cysts in both kidneys that are detectable by MRI, this means that more than 36,000 cysts are actually present, and MRI detects less than 2% of the total cysts present.
Although microcysts are too small to contribute much to total kidney volume, they can interfere with kidney function. Microcysts can reduce GFR in two major ways: by compressing microvasculature, tubules, and glomeruli in the cortex; or by blocking the drainage of multiple upstream nephrons when they form in or block medullary collecting ducts.52 Although the growth rates of microcysts less than 1 mm in size have not yet been measured, the adult combined growth rates of the renal cyst component is approximately 12% per year, with yearly individual cyst growth rates up to 71%, and with fetal cyst growth rates even higher for cysts larger than 7.0 mm.53 Before and during an accelerated growth period, microcysts may be susceptible to certain therapies that could first improve GFR and only later change measurable total kidney volume by slowing microcyst progression to macrocysts either directly or through specialized cells that may be sensitive to rapamycin.
CURRENT MANAGEMENT OF ADPKD
Blood pressure control is essential—but too low is not good. For adult patients with hypertension caused by ADPKD, an acceptable blood pressure range is 120–130/70–80 mm Hg. However, further information from recently published blood pressure guidelines54 and the results of the Halt Progression of Polycystic Kidney Disease (HALT-PKD) study to be reported in late 201455 may provide more precise ranges for blood pressure control in ADPKD.
Although substantial experimental evidence exists for the benefits of inhibiting the up-regulation of the renin-angiotensin-aldosterone system in ADPKD, clinical proof is not yet available to confirm that angiotensin-converting enzyme (ACE) inhibitors or angiotensin receptor blockers (ARBs) are preferred therapy.55 This may be determined by results of the HALT-PKD study, due for release in late 2014.55
Controlling blood pressure should be done with caution. Patients with low GFRs whose blood pressure is too low tend to have a more rapid decline of GFR, as suggested in the Modification of Diet in Renal Disease (MDRD) study in 1995.56
Experimental evidence suggests that avoiding calcium channel blockers may be advisable. Yamaguchi et al34 found that calcium channel blockers worsen the calcium transport defect and convert tubular epithelial cells to a proliferative phenotype.34
High fluid intake (2,500–3,000 mL/day), because it suppresses vasopressin, may be useful if permitted by several factors such as the patient’s cardiopulmonary and renal and electrolyte status, other medications, and diet.31 The reader is referred to a detailed description of the precautions necessary when prescribing high water intake.31 Patients should have their fluid intake managed by a physician and their renal function and serum sodium and electrolytes monitored regularly in order to avoid hyponatremia. Severe hyponatremia has occurred in patients with ADPKD and impaired kidney function who drank excessive quantities of water. Cardiac and pulmonary complications from excessive fluid intake are also possible, especially with a low GFR and compromised cardiac function.
A low-sodium diet, if not a contributing factor in hyponatremia, can be used under physician direction in the management of hypertension as well as in the prevention of calcium oxalate kidney stones.
Caffeine should be avoided because it may interfere with the activity of the phosphodiesterase that is necessary for the catabolism of cAMP to 5′AMP.
A low-protein diet is of unproven benefit,56 but it is prudent to avoid high protein intake.57
Complications such as bleeding (into or from cysts), infection (urinary tract, kidney cysts, and liver cysts), kidney stones, and urinary tract obstruction should be treated promptly and may require hospitalization.
Regular symptom reviews and physical examinations need to be performed with nonrenal concerns also in mind, such as intracranial aneurysms and cardiac valve lesions.11,58
Formal genetic counseling and molecular testing are becoming more frequently indicated as more complex inheritance patterns arise.6–8,59
Renal replacement therapy in the form of dialysis or transplantation is usually available for the patient when end-stage renal disease occurs. In the largest study thus far, ADPKD patient survival with a kidney transplant was similar to that of patients without ADPKD (about 93% at 5 years), and from 5 years to 15 years death-censored graft survival was actually better.60 Thromboembolic events are more frequent after transplantation,27,60 but they may also occur before transplantation from a massive right kidney compressing the iliac vein or the inferior vena cava, or both, leading to thrombus formation.26
Investigational as well as standard drug studies have intensified. Results from a large randomized study in approximately 1,000 adult ADPKD patients that evaluated over 6 to 8 years the effects of ACE inhibition with or without ARB treatment of hypertension, at both usual and lower blood pressure ranges in those with good renal function, are expected in late 2014.55 Outcomes from a few small clinical studies, eg, one with long-acting somatostatin31,61 and one using low-dose rapamycin48 in adults with ADPKD, will require confirmation in large randomized placebo-controlled clinical studies. In a new 3-year randomized placebo-controlled study of 91 children and young adults (ages 8 to 22) with ADPKD and essentially normal renal function who continued treatment with lisinopril, the addition of pravastatin (20 mg or 40 mg daily based on age) resulted in a significant reduction in the number of patients (46% vs 68%, respectively, P = .03) experiencing a greater than 20% change (increase) in height-adjusted total kidney volume.62 Change in GFR was not reported,62 but an earlier 4-week study in 10 patients treated with simvastatin did show an increase in renal blood flow and GFR.63 Numerous other agents that lack human studies include some described in older experimental work (eg, amiloride,31,64 citrate31,65) and many others from a growing list of potential therapeutic targets.31,66–73 It must be emphasized that there is no FDA-approved medication specifically for the treatment of ADPKD.
Future specific treatments of ADPKD may also involve minimally toxic doses of combination or sequential therapy directed at precystic and then both micro- and macrocystic/cystic fluid expansion aspects of ADPKD.48,74 Unfortunately, at the present time there is no specific FDA-approved therapy for ADPKD.
Autosomal dominant polycystic kidney disease (ADPKD) is the most common inherited renal disease, has an estimated prevalence of 1:400 to 1:1,000 live births in the United States, and occurs worldwide.1,2 There are about 700,000 people living with it in the United States, and about 6,000 new cases arise annually. It accounts for nearly 5% of all patients with end-stage renal disease in the United States.3
This paper will offer an overview of the pathogenesis of renal cysts, review some of the clinical aspects of ADPKD including diagnosis and management of complications, and discuss recent drug trials and current management.
TWO TYPES—PKD1 IS MORE COMMON AND PROGRESSES MORE RAPIDLY
Two major forms of ADPKD are recognized and can usually be determined by genetic testing: PKD1, accounting for about 85% of cases, and PKD2, accounting for 15%.
The gene locus for PKD1 is on the short arm of the 16th chromosome (16p13.3), and its glycoprotein gene product is polycystin 1 (PC1), a large molecule with 4,303 amino acids.2 PC1 has a long N-terminal extracellular tail that can function as a mechanosensor. Disease progression is much faster with PKD1, and end-stage renal disease usually occurs before age 56.4
In PKD2, the gene locus is on the long arm of the fourth chromosome (4q21–23), and has a smaller glycoprotein gene product, polycystin 2 (PC2), that plays a role in calcium transport. The disease course of PKD2 tends to be slower. End-stage renal disease might not develop in the patient’s lifetime, since it typically develops when the patient is more than 70 years old.4
Although the growth rate of renal cysts is similar between the two types, patients with PKD1 develop about twice as many cysts as those with PDK2, and their cyst development starts at a younger age.5
Typically, patients have a clear phenotype and a positive family history, but in about 10% of possible ADPKD cases, there is no family history of ADPKD. Genetic variations such as incompletely penetrant PKD1 alleles,6 hypomorphic alleles,7 and trans-heterozygous mutations8 account for at least some of these cases.
IMAGING CRITERIA HAVE BROADENED
Ultrasonographic criteria for the diagnosis of ADPKD that were published in 1994 were based on patients who had a family history of PKD1.9 The criteria have since been modified (the “unified criteria”) to include patients with a family history of PKD2 who begin cyst development at a later age and with lower numbers.10 For patients ages 30 to 39, a previously difficult diagnostic group, the criterion for the minimum number of cysts visible on ultrasonography changed from four to three, improving the sensitivity of detecting disease from approximately 76% to approximately 95% (Table 1).9,10 It is important to note that these criteria apply only to patients “at risk,” ie, with a positive family history of ADPKD.
Computed tomography (CT) and magnetic resonance imaging (MRI) classically show bilaterally enlarged multicystic kidneys, though variations can be seen.
DISEASE CAN PRESENT IN MYRIAD WAYS
Although cystic kidney disease is the basic underlying problem, undiagnosed patients may present with a variety of symptoms caused by other manifestations of ADPKD (Table 2).
Hypertension is the most common presentation, occurring in about 50% of patients ages 20 to 34, and essentially 100% of those with end-stage renal disease.11 It is associated with up-regulation of the renin-angiotensin-aldosterone system.
Pain is typically located in the abdomen, flank, or back and can occur in a localized or diffuse manner. Early abdominal distress is often simply described as “fullness.” Localized pain is usually caused by bleeding into or rupture of a cyst, renal stones, or infection.12 Because renal cysts are noncommunicating, bleeding can occur into a cyst and cause pain without gross hematuria. Compression by greatly enlarged kidneys, liver, or both can cause a variety of gastrointestinal symptoms such as reflux esophagitis and varying degrees of constipation. Diffuse pain is often musculoskeletal and related to exaggerated lordosis from increasing abdominal size due to enlarging cystic kidneys and sometimes liver.12 In carefully selected cases, cyst aspiration may be helpful.11
Although renal carcinomas are rare and not more frequent than in the general population, they can occur at an earlier age and with constitutional symptoms.11
Urinary tract infections are increased in frequency. A patient may have a simple urinary tract infection that is cured with the appropriate antibiotic. However, a urinary tract infection repeatedly recurring with the same organism is a strong clue that an infected cyst is the source and requires more intensive treatment with the appropriate cyst-penetrating antibiotic. On the other hand, because cysts are noncommunicating, an infected cyst might be present despite a negative urine culture.
Identifying infected cysts can be a challenge with conventional imaging techniques, but combined positron emission tomography and CT (PET/CT) can be a valuable though expensive diagnostic tool to identify an infected kidney or liver cyst, or to identify an unsuspected source of the pain and infection.13
Jouret et al13 evaluated 27 PET/CT scans performed in 24 patients with ADPKD and suspicion of an abdominal infection. Patients were deemed to have probable cyst infection if they met all of the following criteria: temperature more than 38°C for longer than 3 days, loin or liver tenderness, plasma C-reactive protein level greater than 5 mg/dL, and no evidence of intracystic bleeding on CT. Patients with only two or three of these criteria were classified as having fever of unknown origin. Diagnosis of cyst infection was confirmed by cyst fluid analysis.
PET/CT identified a kidney or liver cyst infection in 85% of 13 infectious events in 11 patients who met all the criteria for probable cyst infection; CT alone contributed to the diagnosis in only one patient.13 In those with fever of unknown origin, PET/CT identified a source of infection in 64% of 14 events in 13 patients: two infected renal cysts, as well as one patient each with other infections that would be difficult to diagnose clinically, ie, small bowel diverticulitis, psoas abscess, diverticulitis of the right colon, pyelonephritis in a transplanted kidney, infected abdominal aortic aneurysm, prostatitis, colitis, and Helicobacter pylori gastritis. Results of PET/CT were negative in five patients with intracystic bleeding.
Kidney stones occur in 20% to 36% of patients.11,14 Uric acid stones occur at almost the same frequency as calcium oxalate stones.
Chronic kidney disease not previously diagnosed may be the presenting condition in a small percentage of patients, sometimes those in whom much earlier hypertension was not fully evaluated. ADPKD is typically not associated with significant proteinuria (eg, nephrotic range), and the presence of heavy proteinuria usually indicates the presence of a superimposed primary glomerulopathy.15
Cysts in other locations. By MRI, liver cysts are present in 58% of patients ages 15 to 24, rising to 94% in those ages 35 to 46.11 Because liver cysts are estrogen-dependent, they are more prominent in women. A small percentage of patients develop cysts in the pancreas (5%), arachnoid membranes (8%), and seminal vesicles (40% of men with ADPKD).11
Cardiovascular abnormalities occur in almost one-third of patients with ADPKD, usually as mitral and aortic valve abnormalities.16 Aneurysms of the aortic root and abdominal aorta can also occur, in addition to intracranial aneurysms (see below).17
Intracranial aneurysms are not uncommon, and size usually determines their risk.
Intracranial aneurysms are strongly influenced by family history: 16% of ADPKD patients with a family history of intracranial aneurysm also develop them, compared with 5% to 6% of patients with no family history.11 The anterior cerebral circulation is involved in about 80% of cases. A sentinel or sudden “thunderclap” headache is a classic presentation that may precede full-blown rupture in about 17% of cases.18 Patients who rupture an intracranial aneurysm have a mean age of 39, usually have normal renal function, and can be normotensive.11
For patients with no history of subarachnoid hemorrhage, the 5-year cumulative rupture rates for patients with aneurysms located in the internal carotid artery, anterior communicating or anterior cerebral artery, or middle cerebral artery were 0% for aneurysms less than 7 mm, 2.6% for those 7 to 12 mm, 14.5% for those 13 to 24 mm, and 40% for those 25 mm or larger, with higher rates for the same sizes in the posterior circulation.11
In patients without symptoms, size is correlated with risk of rupture: less than 4 mm is usually associated with very low risk, 4 to less than 7 mm with moderate risk, and 7 mm or more with increasing risk. An aneurysm larger than 10 mm is associated with roughly a 1% risk of rupture per year.19
Irazabal et al20 retrospectively studied 407 patients with ADPKD who were screened for intracranial aneurysm. Saccular aneurysms were detected in 45 patients; most were small (median diameter 3.5 mm). During cumulative imaging follow-up of 243 years, only one new intracranial aneurysm was detected (increasing from 2 to 4.4 mm over 144 months) and two previously identified aneurysms grew (one increasing 4.5 to 5.9 mm over 69 months and the other 4.7 to 6.2 mm over 184 months). No change occurred in 28 patients. Seven patients were lost to follow-up, however. During cumulative clinical follow-up of 316 years, no aneurysm ruptured. Two patients were lost to follow-up, three had surgical clipping, and five died of unrelated causes. The authors concluded that presymptomatic intracranial aneurysms are usually small, and that growth and rupture risks are no higher than for unruptured intracranial aneurysms in the general population. A 2014 study also suggests a conservative approach for managing intracranial aneurysm in the general population.21
In asymptomatic ADPKD patients, it is reasonable to reserve screening for those with a positive family history of intracranial aneurysm or subarachnoid hemorrhage, those with a previous ruptured aneurysm, those in high-risk professions (eg, pilots), and for patients prior to anticoagulant therapy or major surgery possibly associated with hemodynamic instability.11,22 Certain extremely anxious patients might also need to be studied. Screening can be performed with magnetic resonance angiography without gadolinium contrast. It is prudent to have patients with an intracranial aneurysm thoroughly evaluated by an experienced neurosurgeon with continued follow-up.
PROGRESSION OF ADPKD
The Consortium for Radiologic Imaging Studies of Polycystic Kidney Disease (CRISP) study23 evaluated 241 patients with ADPKD (ages 15 to 46) by measuring the annual rate of change in total kidney volume, total cyst volume, and iothalamate glomerular filtration rate (GFR) over 3 years. The annual increase in total kidney volume averaged 5.3%,23 though the reported range with various imaging techniques is from 4% to 12.8% in adults.24 This study focused on macrocystic disease, ie, cysts that are visible by MRI and measurably increase total kidney volume. Although larger total kidney volume at baseline generally predicted a more rapid decline in GFR, there were wide and overlapping variations in yearly GFR declines within and among different total-kidney-volume groups.23
SPECIAL CLINICAL PROBLEMS IN ADPKD
Case 1: A man with ADPKD develops new and increasing proteinuria
A 55-year-old man with ADPKD and stage 3 chronic kidney disease developed new and increasing proteinuria, rising to 5,500 mg per 24 hours. What is the most likely explanation?
- Rapidly progressive renal failure with increasing proteinuria in ADPKD
- Bilateral renal vein thromboses because of cyst compression
- Malignant hypertension with bilateral renal artery compression
- Superimposed primary glomerulopathy
- Multiple infected renal cysts with pyonephrosis
Answer: Superimposed primary glomerulopathy.
ADPKD (similar to uncomplicated obstructive uropathy, pyelonephritis, main renal artery disease, and often cases of interstitial nephritis without secondary glomerular changes) typically does not result in nephrotic-range proteinuria. A superimposed primary glomerulopathy, focal segmental glomerulosclerosis, was the biopsy-proved diagnosis.
At least 21 cases have been reported of AD-PKD with nephrotic-range proteinuria and a renal biopsy showing a primary glomerulopathy, including focal segmental glomerulosclerosis (5 cases), minimal-change disease (5), membranous nephropathy (3), IgA nephropathy (2), and one each of crescentic glomerulonephropathy, diabetic nephropathy, membranoproliferative glomerulonephritis, postinfectious glomerulonephropathy, amyloid glomerulopathy, and mesangioproliferative glomerulopathy.15 Treatment was directed at the primary glomerulopathy, and the outcomes corresponded to the primary diagnosis (eg, with appropriate treatment, three of the five patients with focal segmental glomerulosclerosis progressed to end-stage renal disease, all of the patients with minimal-change disease went into remission, and one of the two cases with IgA nephropathy improved).15
Case 2: A woman with ADPKD and advanced renal failure develops shortness of breath
A 47-year-old woman with very large polycystic kidneys (total kidney volume 7,500 mL; normal range for a single kidney approximately 136–295 mL, mean 196)25 and estimated GFR of 25 mL/min developed new-onset shortness of breath while climbing steps and later even when making a bed. She had no chest pain, cough, or edema. She was sent directly to the emergency department and was admitted and treated; her condition improved, and she was discharged after 6 days. What did she have?
- Presentation of rare cystic pulmonary disease in ADPKD
- Onset of pneumonia with early bacteremia
- Progressive reduction in ventilatory capacity from massive polycystic kidneys and liver elevating both sides of the diaphragm
- Pulmonary emboli from an iliac vein or inferior vena cava source
- Progressive anemia accompanying rapidly worsening stage 4 chronic kidney disease
Answer: She had pulmonary emboli from an iliac vein (right) or inferior vena cava source.
Pulmonary emboli in ADPKD can be caused by thrombi in the inferior vena cava or the iliac or femoral vein because of compression by a massive right polycystic kidney. Four cases were reported at Mayo Clinic,26 three diagnosed by MRI and one with CT. One additional case occurred at Cleveland Clinic. All patients survived after treatment with anticoagulation therapy; early nephrectomy was required in two cases.
Interestingly, following kidney transplantation, the patients at greatest risk for pulmonary emboli are those with ADPKD as their original disease.27
RENAL CYSTS RESULT FROM COMBINED MUTATIONS, INJURY
The germline ADPKD mutation that occurs in one allele of all renal tubular epithelial cells is necessary but not sufficient for cystogenesis.28 One or more additional somatic mutations of the normal allele—the “second hit”—also develop within individual tubular epithelial cells.28,29 These epithelial cells undergo clonal proliferation, resulting in tubular dilatation and cyst formation. Monoclonality of cells in cysts has been documented.
Ischemia-reperfusion injury can be viewed as a “third hit.”30 In PKD1 knockout mice, which at 5 weeks of age normally develop only mild cystic kidney disease, the superimposition of unilateral ischemia-reperfusion injury at 8 weeks caused widespread and rapid cyst formation. It is believed that acute renal injury reactivates developmental signaling pathways within 48 hours that trigger epithelial cell proliferation and then cyst development detectable by MRI 2 weeks later. Although this phenomenon has not been documented in humans, it is a cautionary tale.
CYSTOGENESIS INVOLVES MULTIPLE PATHWAYS
A comprehensive description of pathways leading to renal cyst formation is beyond the scope of this article, and the reader is referred to much more detailed and extensive reviews.2,31 Disturbances in at least three major interconnected pathways promote cystogenesis in renal tubular epithelial cells:
- Normal calcium transport into the endoplasmic reticulum is disrupted by abnormal polycystins on the surface of the primary cilium
- Vasopressin and other stimuli increase the production of cyclic adenosine monophosphate (cAMP)
- The mammalian target of rapamycin (mTOR) proliferative pathway is up-regulated.
DISRUPTION OF CALCIUM TRANSPORT IN THE PRIMARY CILIUM
Primary cilia are nonmotile cellular organelles of varying size, from about 0.25 μm up to about 1 μm.32 Each primary cilium has nine peripheral pairs of microtubules but lacks a centrally located pair that is present in motile cilia. Primary cilia are ubiquitous and have been highly conserved throughout evolution. A single cilium is present on almost all vertebral cells.33
Cilial defects have been identified in autosomal dominant as well as recessive diseases and are known as ciliopathies.33 Although rare in humans, they can affect a broad spectrum of organs other than the kidney, including the eye, liver, and brain.33
Urine flow in a renal tubule is believed to exert mechanical stimulation on the extracellular flagellum-like N-terminal tail of PC1 that extends from a primary cilium into the urinary space. PC1 in concert with PC2 opens PC2 calcium channels, allowing calcium ions to flow down the microtubules to ryanodine receptors and the basal body.32,33 This leads to local release of calcium ions that regulate cell proliferation.32,34 However, in ADPKD kidneys, PC1 and PC2 molecules are sparse or mutated, resulting in defective calcium transport, increased and unregulated tubular epithelial cell proliferation, and cyst formation.
In a totally different clinical setting, biopsies of human renal transplants that sustained acute tubular necrosis during transplantation reveal that a cilium dramatically elongates in response to injury,35 possibly as a compensatory mechanism to maintain calcium transport in the presence of meager urine flow and to restore the proliferation of tubular epithelial cells in a regulated repair process.
THE ROLE OF VASOPRESSIN AND ACTIVATION OF cAMP
In classic experiments, Wang et al36 cross-bred rats having genetically inherited polycystic kidney disease (actually, autosomal recessive polycystic kidney disease) with Brattleboro rats that completely lack vasopressin. At 10 and 20 weeks of age, the offspring had virtually complete inhibition of cystogenesis because of the absence of vasopressin. However, when vasopressin was restored by exogenous administration continuously for 8 weeks, the animals formed massive renal cysts.
Vasopressin activates cAMP, which then functions as a second messenger in cell signaling. cAMP increases the activation of the protein kinase A (PKA) pathway, which in turn increases downstream activity of the B-raf/ERK pathway. Up-regulation of cAMP and PKA appears to perpetuate activation of canonical Wnt signaling, down-regulate non-canonical Wnt/planar cell polarity signaling, and lead to loss of tubular diameter control, resulting in cyst formation.31 Normally, cAMP is degraded by phosphodiesterase. However, because of the primary cilium calcium transport defect in ADPKD, phosphodiesterase is reduced and cAMP persists.37 In conjunction with the defective primary cilial calcium transport, cAMP exerts a proliferative effect on renal tubular epithelial cells that is opposite to its effect in normal kidneys.31,32 cAMP also up-regulates the cystic fibrosis transmembrane conductance regulator (CFTR) that promotes chloride ion transport. Sodium ions follow the chloride ions, leading to fluid accumulation and cyst enlargement.31
Inhibiting vasopressin by increasing water intake
A simple key mechanism for limiting vasopressin secretion is by sufficient water ingestion. Nagao et al38 found that rats with polycystic kidney disease given water with 5% glucose (resulting in 3.5-fold increased fluid intake compared with rats given tap water) had a 68% reduction in urinary vasopressin and a urine osmolality less than 290 mOsm/kg. The high-water-intake rats had dramatically reduced cystic areas in the kidney and a 28% reduction of kidney-to-body weight ratio vs controls.
In an obvious oversimplification, these findings raised the question of whether a sufficient increase in water intake could be an effective therapy for polycystic kidney disease.39 A pilot clinical study evaluated changes in urine osmolality in eight patients with ADPKD who had normal renal function.40 At baseline, 24-hour urine osmolality was typically elevated to approximately 753 mOsm/kg compared to the plasma at 285 mOsm/kg, indicating that antidiuresis is the usual state. During the 2-week study, urine volume and osmolality were measured, and additional water intake was adjusted in order to achieve a urine osmolality goal of 285 ± 45 mOsm/kg. These adjustments resulted in water intake that appeared to be in the range of 2,400 to 3,000 mL per 24 hours. The major limitations of the study were that it was very short term, and there was no opportunity to measure changes in total kidney volume or estimated GFR.
In a recent preliminary report from Japan, high water intake (2,500–3,000 mL daily) in 18 ADPKD patients was compared over 12 months with ad libitum water intake in 14 ADPKD controls (clinicaltrials.gov NCT 01348505). There was no statistically significant change in total kidney volume or cystatin-estimated GFR in those on high water intake, but serious defects in study design (patients in the high water intake group were allowed to decrease their intake if it was causing them difficulty, and patients in the ad libitum water intake group had no measurement of their actual water intake) prevent any conclusions because there was no evidence that the groups were different from one another with respect to the key element of the study, namely, water intake.
Blocking the vasopressin receptor slows disease progression
Using another approach, Gattone et al41 inhibited the effect of vasopressin by blocking the vasopressin 2 receptor (V2R) in mouse and rat models of polycystic kidney disease, using an experimental drug, OPC31260. The drug halted disease progression and, in one situation, appeared to cause regression of established disease. As noted by Torres and Harris,31 even though both increased water intake and V2R antagonists decrease cAMP in the distal tubules and collecting ducts, circulating levels of vasopressin are decreased by increased water intake but increased by V2R antagonists.
After these remarkable results in animal models, clinical trials of the V2R antagonist tolvaptan were conducted in patients with ADPKD. In the Tolvaptan Efficacy and Safety in Management of Autosomal Dominant Polycystic Kidney Disease and Its Outcomes 3:4 study,42 1,445 adults (ages 18 to 50) with ADPKD in 133 centers worldwide were randomized to receive either tolvaptan or placebo for 3 years. Key inclusion criteria included good renal function (estimated GFR ≥ 60 mL/min) and total kidney volume of at least 750 mL (mean 1,700 mL) as measured by MRI. Tolvaptan was titrated to the highest tolerated twice-daily dose (average total of 95 mg/day). All patients were advised to maintain good hydration and to avoid thirst by drinking a glass of water after each urination. Unfortunately, neither water intake nor urine output was measured.
The primary end point was the annual rate of change in total kidney volume, with secondary end points of clinical progression (worsening kidney function, pain, hypertension, albuminuria), and rate of decline in kidney function as measured by the slope of the reciprocal of serum creatinine.42
Patients in the tolvaptan arm had a slower annual increase in total kidney volume than controls (2.8% vs 5.5%, respectively, P < .001) and a slower annual decline in renal function (−2.61 vs −3.81 mg/mL−1, respectively, P < .001).42 More participants in the treatment group withdrew than in the placebo group (23% vs 14%, respectively).
Adverse events occurred more frequently with tolvaptan.42 Liver enzyme elevations of greater than three times the upper limit of normal occurred in 4.4% of patients in the treatment group, leading to a drug warning issued in January 2013. As expected, side effects related to diuresis (urinary frequency, nocturia, polyuria, and thirst) were more frequent in the treatment group, occurring in up to 55% of participants.
The authors noted, “Although maintaining hydration helped ensure that the blinding in the study was maintained, the suppression of vasopressin release in the placebo group may have led to an underestimation of the beneficial effect of tolvaptan and may account for the lower rates of kidney growth observed in the placebo group.”42
In 2013, the US Food and Drug Administration (FDA) denied a new drug application for tolvaptan as a treatment for ADPKD.
THE mTOR PATHWAY IS UP-REGULATED
The mTOR pathway that plays a major role in cell growth and proliferation includes interaction of the cytoplasmic tail of polycystin 1 with tuberin.43 Activation products of mTOR, including phospho-S6K, have been found in tubular epithelial cells lining cysts of ADPKD kidneys but not in normal kidneys.43 Mutant mice with polycystic disease had phospho-S6K in tubular epithelial cells of cysts, whereas those treated with the mTOR inhibitor rapamycin did not.43 But subsequent studies have shown that only a low level of mTOR activation is present in 65% to 70% of ADPKD cysts.44
Two major studies of the treatment of ADPKD with rapamycin that were published contemporaneously in 2010 failed to demonstrate any significant benefit with mTOR inhibitor treatment.45,46
Serra et al45 conducted an 18-month, open-label trial of 100 ADPKD patients ages 18 to 40 with an estimated GFR (eGFR) of at least 70 mL/min. Patients were randomized to receive rapamycin, given as sirolimus 2 mg per day, or standard care. The primary end point was the reduction in the growth rate of total kidney volume, measured by MRI. Secondary end points were eGFR and protein excretion (albumin-creatinine ratio). No significant difference was found in total kidney volume, but a nonsignificant stabilization of eGFR was noted.
Walz et al46 in a 2-year, multicenter, double-blind trial, randomized 433 patients (mean age 44; mean eGFR 54.5 mL/min) to treatment with either the short-acting mTOR inhibitor everolimus (2.5 mg twice daily) or placebo. Although patients in the treatment group had less of an increase in total kidney volume (significant at 1 year but not at 2 years), they tended to show a decline in eGFR. But further analysis showed that the only patients who had a reduction in eGFR were males who already had impaired kidney function at baseline.47
In a pilot study, 30 patients with ADPKD (mean age 49) were randomized to one of three therapies:
- Low-dose rapamycin (trough blood level 2–5 ng/mL)
- Standard-dose rapamycin (trough blood level > 5–8 ng/mL)
- Standard care without rapamycin.48
In contrast to other studies, the primary end point was the change in iothalamate GFR at 12 months, with change in total kidney volume being a secondary end point.
At 12 months, with 26 patients completing the study, the low-dose rapamycin group (n = 9) had a significant increase in iothalamate GFR of 7.7 ± 12.5 mL/min/1.73 m2, whereas the standard-dose rapamycin group (n = 8) had a nonsignificant increase of 1.6 ± 12.1 mL/min/1.73 m2, and the no-rapamycin group (n = 9) had a fall in iothalamate GFR of 11.2 ± 9.1 mL/min/1.73 m2 (P = .005 for low-dose vs no rapamycin; P = .07 for standard-dose vs no rapamycin; P = .52 for low-dose vs standard-dose rapamycin; and P = .002 for combined low-dose and standard-dose rapamycin vs no rapamycin.).48 These differences were observed despite there being no significant change in total kidney volume in any of the groups. Patients on low-dose rapamycin had fewer adverse effects than those on standard dose and were more often able to continue therapy for the entire study. This, and the use of iothalamate GFR rather than eGFR to measure GFR, are believed to be the main reasons that low-dose effects were more pronounced than those with standard doses. One may speculate that rapamycin may have its effect on microcysts and cystogenic cells, resulting in stabilization of or improvement in renal function without detectable slowing in total kidney volume enlargement. Mechanisms for this possibility involve new concepts, as discussed below.
NEW CONCEPTS
Specialized cells also promote renal cyst formation
Specialized cells that promote cyst formation have been identified by Karihaloo et al49 in a mouse model of polycystic kidney disease. In this model, alternatively activated macrophages homed to cystic areas and promoted cyst growth. These findings suggested that interrupting the homing and proliferative signals of macrophages could be a therapeutic target for ADPKD. Although rapamycin can suppress macrophage proliferation by macrophage colony-stimulating factor and inhibit macrophage function,50 alternatively activated macrophages have not been specifically studied for rapamycin responsiveness.
More promising is evidence that CD133+ progenitor cells from human ADPKD kidneys—but not from normal human kidneys—form cysts in vitro and in severe combined immunodeficient mouse models.51 Treatment with rapamycin decreased proliferation of the de-differentiated CD133+ cells from ADPKD patients and reduced cystogenesis.51
Visible cysts are the tip of the iceberg
Using ADPKD nephrectomy specimens from eight patients, Grantham et al52 compared cyst counts by MRI and by histology and found that for every renal cyst detected by MRI, about 62 smaller cysts (< 0.9 mm) are present in the kidney. For a typical patient having an average of 587 cysts in both kidneys that are detectable by MRI, this means that more than 36,000 cysts are actually present, and MRI detects less than 2% of the total cysts present.
Although microcysts are too small to contribute much to total kidney volume, they can interfere with kidney function. Microcysts can reduce GFR in two major ways: by compressing microvasculature, tubules, and glomeruli in the cortex; or by blocking the drainage of multiple upstream nephrons when they form in or block medullary collecting ducts.52 Although the growth rates of microcysts less than 1 mm in size have not yet been measured, the adult combined growth rates of the renal cyst component is approximately 12% per year, with yearly individual cyst growth rates up to 71%, and with fetal cyst growth rates even higher for cysts larger than 7.0 mm.53 Before and during an accelerated growth period, microcysts may be susceptible to certain therapies that could first improve GFR and only later change measurable total kidney volume by slowing microcyst progression to macrocysts either directly or through specialized cells that may be sensitive to rapamycin.
CURRENT MANAGEMENT OF ADPKD
Blood pressure control is essential—but too low is not good. For adult patients with hypertension caused by ADPKD, an acceptable blood pressure range is 120–130/70–80 mm Hg. However, further information from recently published blood pressure guidelines54 and the results of the Halt Progression of Polycystic Kidney Disease (HALT-PKD) study to be reported in late 201455 may provide more precise ranges for blood pressure control in ADPKD.
Although substantial experimental evidence exists for the benefits of inhibiting the up-regulation of the renin-angiotensin-aldosterone system in ADPKD, clinical proof is not yet available to confirm that angiotensin-converting enzyme (ACE) inhibitors or angiotensin receptor blockers (ARBs) are preferred therapy.55 This may be determined by results of the HALT-PKD study, due for release in late 2014.55
Controlling blood pressure should be done with caution. Patients with low GFRs whose blood pressure is too low tend to have a more rapid decline of GFR, as suggested in the Modification of Diet in Renal Disease (MDRD) study in 1995.56
Experimental evidence suggests that avoiding calcium channel blockers may be advisable. Yamaguchi et al34 found that calcium channel blockers worsen the calcium transport defect and convert tubular epithelial cells to a proliferative phenotype.34
High fluid intake (2,500–3,000 mL/day), because it suppresses vasopressin, may be useful if permitted by several factors such as the patient’s cardiopulmonary and renal and electrolyte status, other medications, and diet.31 The reader is referred to a detailed description of the precautions necessary when prescribing high water intake.31 Patients should have their fluid intake managed by a physician and their renal function and serum sodium and electrolytes monitored regularly in order to avoid hyponatremia. Severe hyponatremia has occurred in patients with ADPKD and impaired kidney function who drank excessive quantities of water. Cardiac and pulmonary complications from excessive fluid intake are also possible, especially with a low GFR and compromised cardiac function.
A low-sodium diet, if not a contributing factor in hyponatremia, can be used under physician direction in the management of hypertension as well as in the prevention of calcium oxalate kidney stones.
Caffeine should be avoided because it may interfere with the activity of the phosphodiesterase that is necessary for the catabolism of cAMP to 5′AMP.
A low-protein diet is of unproven benefit,56 but it is prudent to avoid high protein intake.57
Complications such as bleeding (into or from cysts), infection (urinary tract, kidney cysts, and liver cysts), kidney stones, and urinary tract obstruction should be treated promptly and may require hospitalization.
Regular symptom reviews and physical examinations need to be performed with nonrenal concerns also in mind, such as intracranial aneurysms and cardiac valve lesions.11,58
Formal genetic counseling and molecular testing are becoming more frequently indicated as more complex inheritance patterns arise.6–8,59
Renal replacement therapy in the form of dialysis or transplantation is usually available for the patient when end-stage renal disease occurs. In the largest study thus far, ADPKD patient survival with a kidney transplant was similar to that of patients without ADPKD (about 93% at 5 years), and from 5 years to 15 years death-censored graft survival was actually better.60 Thromboembolic events are more frequent after transplantation,27,60 but they may also occur before transplantation from a massive right kidney compressing the iliac vein or the inferior vena cava, or both, leading to thrombus formation.26
Investigational as well as standard drug studies have intensified. Results from a large randomized study in approximately 1,000 adult ADPKD patients that evaluated over 6 to 8 years the effects of ACE inhibition with or without ARB treatment of hypertension, at both usual and lower blood pressure ranges in those with good renal function, are expected in late 2014.55 Outcomes from a few small clinical studies, eg, one with long-acting somatostatin31,61 and one using low-dose rapamycin48 in adults with ADPKD, will require confirmation in large randomized placebo-controlled clinical studies. In a new 3-year randomized placebo-controlled study of 91 children and young adults (ages 8 to 22) with ADPKD and essentially normal renal function who continued treatment with lisinopril, the addition of pravastatin (20 mg or 40 mg daily based on age) resulted in a significant reduction in the number of patients (46% vs 68%, respectively, P = .03) experiencing a greater than 20% change (increase) in height-adjusted total kidney volume.62 Change in GFR was not reported,62 but an earlier 4-week study in 10 patients treated with simvastatin did show an increase in renal blood flow and GFR.63 Numerous other agents that lack human studies include some described in older experimental work (eg, amiloride,31,64 citrate31,65) and many others from a growing list of potential therapeutic targets.31,66–73 It must be emphasized that there is no FDA-approved medication specifically for the treatment of ADPKD.
Future specific treatments of ADPKD may also involve minimally toxic doses of combination or sequential therapy directed at precystic and then both micro- and macrocystic/cystic fluid expansion aspects of ADPKD.48,74 Unfortunately, at the present time there is no specific FDA-approved therapy for ADPKD.
- Torres VE, Harris PC. Mechanisms of disease: autosomal dominant and recessive polycystic kidney diseases. Nat Clin Pract Nephrol 2006; 2:40–55.
- Torres VE, Harris PC. Autosomal dominant polycystic kidney disease: the last 3 years. Kidney Int 2009; 76:149–168.
- United States Renal Data System. 2013 atlas of CKD & ESRD. Volume 2 - atlas ESRD:172. www.usrds.org/atlas.aspx. Accessed June 4, 2014.
- Barua M, Cil O, Paerson AD, et al. Family history of renal disease severity predicts the mutated gene in ADPKD. J Am Soc Nephrol 2009, 20:1833–1838.
- Harris PC, Bae KT, Rossetti S, et al. Cyst number but not the rate of cystic growth is associated with the mutated gene in autosomal dominant polycystic kidney disease. J Am Soc Nephrol 2006; 17:3013–3019.
- Vujic M, Heyer CM, Ars E, et al. Incompletely penetrant PKD1 alleles mimic the renal manifestations of ARPKD. J Am Soc Nephrol 2010; 21:1097–1102.
- Harris PC. What is the role of somatic mutation in autosomal dominant polycystic kidney disease? J Am Soc Nephrol 2010; 21:1073–1076.
- Watnick T, He N, Wang K, et al. Mutations of PKD1 in ADPKD2 cysts suggest a pathogenic effect of trans-heterozygous mutations. Nat Genet 2000; 25:143–144.
- Ravine D, Gibson RN, Walker RG, Sheffield LJ, Kincaid-Smith P, Danks DM. Evaluation of ultrasonographic diagnostic criteria for autosomal dominant polycystic kidney disease 1. Lancet 1994; 343:824–827.
- Pei Y, Obaji J, Dupuis A, et al. Unified criteria for ultrasonographic diagnosis of ADPKD. J Am Soc Nephrol 2009; 20:205–212.
- Torres VE, Harris PC, Pirson Y. Autosomal dominant polycystic kidney disease. Lancet 2007; 369:1287–1301.
- Bajwa ZH, Sial KA, Malik AB, Steinman TI. Pain patterns in patients with polycystic kidney disease. Kidney Int 2004; 66:1561–1569.
- Jouret F, Lhommel R, Beguin C, et al. Positron-emission computed tomography in cyst infection diagnosis in patients with autosomal dominant polycystic kidney disease. Clin J Am Soc Nephrol 2011; 6:1644–1650.
- Nishiura JL, Neves RF, Eloi SR, Cintra SM, Ajzen SA, Heilberg IP. Evaluation of nephrolithiasis in autosomal dominant polycystic kidney disease patients. Clin J Am Soc Nephrol 2009; 4:838–844.
- Hiura T, Yamazaki H, Saeki T, et al. Nephrotic syndrome and IgA nephropathy in polycystic kidney disease. Clin Exp Nephrol 2006; 10:136–139.
- Hossack KF, Leddy CL, Johnson AM, Schrier RW, Gabow PA. Echocardiographic findings in autosomal dominant polycystic kidney disease. N Engl J Med 1988; 319:907–912.
- Rossetti S, Chauveau D, Kubly V, et al. Association of mutation position in polycystic kidney disease 1 (PKD1) gene and development of a vascular phenotype. Lancet 2003; 361:2196–2201.
- Linn FH, Wijdicks EF, van der Graaf Y, Weerdesteyn-van Vliet FA, Bartelds AI, van Gijn J. Prospective study of sentinel headache in aneurismal subarachnoid haemorrhage. Lancet 1994; 344:590–593.
- Belz MM, Fick-Brosnahan GM, Hughes RL, et al. Recurrence of intracranial aneurysms in autosomal-dominant polycystic kidney disease. Kidney Int 2003; 63:1824–1830.
- Irazabal MV, Huston J, Kubly V, et al. Extended follow-up of unruptured intracranial aneurysms detected by presymptomatic screening in patients with autosomal dominant polycystic kidney disease. Clin J Am Soc Nephrol 2011; 6:1274–1285.
- Salman A-S, White PM, Counsell CE, et al; Scottish Audit of Intracranial Vascular Malformations Collaborators. Outcome after conservative management or intervention for unruptured brain arteriovenous malformations. JAMA 2014; 311:1661–1669.
- Vijay A, Vijay A, Pankaj P. Autosomal dominant polycystic kidney disease: a comprehensive review. Nephrourol Mon 2010; 2:172–192.
- Grantham JJ, Torres VE, Chapman AB, et al; CRISP Investigators. Volume progression in polycystic kidney disease. N Engl J Med 2006; 354:2122–2130.
- Bae KT, Grantham JJ. Imaging for the prognosis of autosomal dominant polycystic kidney disease. Nat Rev Nephrol 2010; 6:96–106.
- van den Dool SW, Wasser NM, de Fijter JW, Hoekstra J, van der Geest RJ. Functional renal volume: quantitative analysis at gadolinium-enhanced MR angiography—feasibility study in healthy potential kidney donors. Radiology 2005; 236:189–195.
- O’Sullivan DA, Torres VE, Heit JA, Liggett S, King BF. Compression of the inferior vena cava by right renal cysts: an unusual cause of IVC and/or iliofemoral thrombosis with pulmonary embolism in autosomal dominant polycystic kidney disease. Clin Nephrol 1998; 49:332–334.
- Tveit DP, Hypolite I, Bucci J, et al. Risk factors for hospitalizations resulting from pulmonary embolism after renal transplantation in the United States. J Nephrol 2001; 14:361–368.
- Pei Y. A “two-hit” model of cystogenesis in autosomal dominant polycystic kidney disease? Trends Mol Med 2001; 7:151–156.
- Qian F, Germino GG. “Mistakes happen”: somatic mutation and disease. Am J Hum Genet 1997; 61:1000–1005.
- Takakura A, Contrino L, Zhou X, et al. Renal injury is a third hit promoting rapid development of adult polycystic kidney disease. Hum Mol Genet 2009; 18:2523–2531.
- Torres VE, Harris PC. Strategies targeting cAMP signaling in the treatment of polycystic kidney disease. J Am Soc Nephrol 2014; 25:18–32.
- Nauli SM, Alenghat FJ, Luo Y, et al. Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat Genet 2003; 33:129–137.
- Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N Engl J Med 2011; 364:1533–1543.
- Yamaguchi T, Wallace DP, Magenheimer BS, Hempson SJ, Grantham JJ, Calvet JP. Calcium restriction allows cAMP activation of the B-Raf/ERK pathway, switching cells to a cAMP-dependent growth-stimulated phenotype. J Biol Chem 2004; 279:40419–40430.
- Verghese E, Ricardo SD, Weidenfeld R, et al. Renal primary cilia lengthen after acute tubular necrosis. J Am Soc Nephrol 2009; 20:2147–2153.
- Wang X, Wu Y, Ward CJ, Harris PC, Torres VE. Vasopressin directly regulates cyst growth in polycystic kidney disease. J Am Soc Nephrol 2008; 19:102–108.
- Torres VE. Cyclic AMP, at the hub of the cystic cycle. Kidney Int 2004; 66:1283–1285.
- Nagao S, Nishii K, Katsuyama M, et al. Increased water intake decreases progression of polycystic kidney disease in the PCK rat. J Am Soc Nephrol 2006; 17:2220–2227.
- Grantham JJ. Therapy for polycystic kidney disease? It’s water, stupid! J Am Soc Nephrol 2008; 19:1–7.
- Wang CJ, Creed C, Winklhofer FT, Grantham JJ. Water prescription in autosomal dominant polycystic kidney disease: a pilot study. Clin J Am Soc Nephrol 2011; 6:192–197.
- Gattone VH, Wang X, Harris PC, Torres VE. Inhibition of renal cystic disease development and progression by a vasopressin V2 receptor antagonist. Nat Med 2003; 9:1323–1326.
- Torres VE, Chapman AB, Devuyst O, et al; TEMPO 3:4 Trial Investigators. Tolvaptan in patients with autosomal dominant polycystic kidney disease. N Engl J Med 2012; 367:2407–2418.
- Shillingford JM, Murcia NS, Larson CH, et al. The mTOR pathway is regulated by polycystin-1, and its inhibition reverses renal cystogenesis in polycystic kidney disease. Proc Natl Acad Sci U S A 2006; 103:5466–5471.
- Hartman TR, Liu D, Zilfou JT, et al. The tuberous sclerosis proteins regulate formation of the primary cilium via a rapamycin-insensitive and polycystin 1-independent pathway. Hum Mol Genet 2009; 18:161–163.
- Serra AL, Poster D, Kistler AD, et al. Sirolimus and kidney growth in autosomal dominant polycystic kidney disease. N Engl J Med 2010; 363:820–829.
- Walz G, Budde K, Mannaa M, et al. Everolimus in patients with autosomal dominant polycystic kidney disease. N Engl J Med 2010; 363:830–840. Errata in: N Engl J Med 2010; 363:1190 and N Engl J Med 2010; 363:1977.
- Walz G, Budde K, Eckardt K-U. mTOR inhibitors and autosomal dominant polycystic kidney disease (correspondence). N Engl J Med 2011; 364:287–288.
- Braun WE, Schold JD, Stephany BR, Spinko RA, Herfs BR. Low dose rapamycin (sirolimus) effects in autosomal dominant polycystic kidney disease: an open-label randomized control pilot study. Clin J Am Soc Nephrol 2014; 9:881–888.
- Karihaloo A, Koraishy F, Huen SC, et al. Macrophages promote cyst growth in polycystic kidney disease. J Am Soc Nephrol 2011; 22:1809–1814.
- Fox R, Nhan TQ, Law GL, Morris DR, Liles WC, Schwartz SM. PSGL-1 and mTOR regulate translation of ROCK-1 and physiological functions of macrophages. EMBO J 2007; 26:505–515. Erratum in: EMBO J 2007; 26:2605.
- Carvalhosa R, Deambrosis I, Carrera P, et al. Cystogenic potential of CD133+ progenitor cells of human polycystic kidneys. J Pathol 2011; 225:129–141.
- Grantham JJ, Mulamalla S, Grantham CJ, et al. Detected renal cysts are tips of the iceberg in adults with ADPKD. Clin J Am Soc Nephrol 2012; 7:1087–1093.
- Grantham JJ, Cook LT, Wetzel LH, Cadnapaphornchai MA, Bae KT. Evidence of extraordinary growth in the progressive enlargement of renal cysts. Clin J Am Soc Nephrol 2010; 5:889–896.
- James PA, Oparil S, Carter BL, et al. 2014 evidence-based guideline for the management of high blood pressure in adults: report from the panel members appointed to the Eighth Joint National Committee (JNC 8). JAMA 2014; 311:507–520.
- Chapman AB, Torres VE, Perrone RD, et al. The HALT polycystic kidney disease trials: design and implementation. Clin J Am Soc Nephrol 2010; 5:102–109.
- Klahr S, Breyer JA, Beck GJ, et al. Dietary protein restriction, blood pressure control, and the progression of polycystic kidney disease. Modification of Diet in Renal Disease Study Group. J Am Soc Nephrol 1995; 5:2037–2047.
- Thilly N. Low-protein diet in chronic kidney disease: from questions of effectiveness to those of feasibility. Nephrol Dial Transplant 2013; 28:2203–2205.
- Luciano RL, Dahl NK. Extra-renal manifestations of autosomal dominant polycystic kidney disease (ADPKD): considerations for routine screening and management. Nephrol Dial Transplant 2014; 29:247–254.
- Harris PC, Rossetti S. Molecular diagnostics for autosomal dominant polycystic kidney disease. Nat Rev Nephrol 2010; 6:197–206.
- Jacquet A, Pallet N, Kessler M, et al. Outcomes of renal transplantation in patients with autosomal dominant polycystic kidney disease: a nationwide longitudinal study. Transpl Int 2011; 24:582–587.
- Ruggenenti P, Remuzzi A, Ondei P, et al. Safety and efficacy of long-acting somatostatin treatment in autosomal-dominant polycystic kidney disease. Kidney Int 2005; 68:206–216.
- Cadnapaphornchai MA, George DM, McFann K, et al. Effect of pravastatin on total kidney volume, left ventricular mass index, and microalbuminuria in pediatric autosomal dominant polycystic kidney disease. Clin J Am Soc Nephrol 2014; 9:889–896.
- van Dijk MA, Kamper AM, van Veen S, Souverjin JH, Blauw GJ. Effect of simvastatin on renal function in autosomal dominant polycystic kidney disease. Nephrol Dial Transplant 2001; 16:2152–2157.
- Grantham JJ, Uchich M, Cragoe EL, et al. Chemical modification of cell proliferation and fluid secretion in renal cysts. Kidney Int 1989; 35:1379–1389.
- Tanner GA. Potassium citrate/citric acid intake improves renal function in rats with polycystic kidney disease. J Am Soc Nephrol 1998; 9:1242–1248.
- Belibi FA, Edelstein CL. Novel targets for the treatment of autosomal dominant polycystic kidney disease. Expert Opin Investig Drugs 2010; 19:315–328.
- Tao Y, Kim J, Yin Y, et al. VEGF receptor inhibition slows the progression of polycystic kidney disease. Kidney Int 2007; 72:1358–1366.
- Terryn S, Ho A, Beauwens R, Devuyst O. Fluid transport and cystogenesis in autosomal dominant polycystic kidney disease. Biochim Biophys Acta 2011; 1812:1314–1321.
- Thiagarajah JR, Verkman AS. CFTR inhibitors for treating diarrheal disease. Clin Pharmacol Ther 2012; 92:287–290.
- Boehn SN, Spahn S, Neudecker S, et al. Inhibition of Comt with tolcapone slows proression of polycystic kidney disease in the more severely affected PKD/Mhm (cy/+) substrain of the Hannover Sprague-Dawley rat. Nephrol Dial Transplant 2013; 28:2045–2058.
- Rees S, Kittikulsuth W, Roos K, Strait KA, Van Hoek A, Kohan DE. Adenylyl cyclase 6 deficiency ameliorates polycystic kidney disease. J Am Soc Nephrol 2014; 25:232–237.
- Buchholz B, Schley G, Faria D, et al. Hypoxia-inducible factor-1a causes renal cyst expansion through calcium-activated chloride secretion. J Am Soc Nephrol 2014; 25:465–474.
- Wallace DP, White C, Savinkova L, et al. Periostin promotes renal cyst growth and interstitial fibrosis in polycystic kidney disease. Kidney Int 2014; 85:845–854.
- Leuenroth SJ, Crews CM. Targeting cyst initiation in ADPKD. J Am Soc Nephrol 2009; 20:1–3.
- Torres VE, Harris PC. Mechanisms of disease: autosomal dominant and recessive polycystic kidney diseases. Nat Clin Pract Nephrol 2006; 2:40–55.
- Torres VE, Harris PC. Autosomal dominant polycystic kidney disease: the last 3 years. Kidney Int 2009; 76:149–168.
- United States Renal Data System. 2013 atlas of CKD & ESRD. Volume 2 - atlas ESRD:172. www.usrds.org/atlas.aspx. Accessed June 4, 2014.
- Barua M, Cil O, Paerson AD, et al. Family history of renal disease severity predicts the mutated gene in ADPKD. J Am Soc Nephrol 2009, 20:1833–1838.
- Harris PC, Bae KT, Rossetti S, et al. Cyst number but not the rate of cystic growth is associated with the mutated gene in autosomal dominant polycystic kidney disease. J Am Soc Nephrol 2006; 17:3013–3019.
- Vujic M, Heyer CM, Ars E, et al. Incompletely penetrant PKD1 alleles mimic the renal manifestations of ARPKD. J Am Soc Nephrol 2010; 21:1097–1102.
- Harris PC. What is the role of somatic mutation in autosomal dominant polycystic kidney disease? J Am Soc Nephrol 2010; 21:1073–1076.
- Watnick T, He N, Wang K, et al. Mutations of PKD1 in ADPKD2 cysts suggest a pathogenic effect of trans-heterozygous mutations. Nat Genet 2000; 25:143–144.
- Ravine D, Gibson RN, Walker RG, Sheffield LJ, Kincaid-Smith P, Danks DM. Evaluation of ultrasonographic diagnostic criteria for autosomal dominant polycystic kidney disease 1. Lancet 1994; 343:824–827.
- Pei Y, Obaji J, Dupuis A, et al. Unified criteria for ultrasonographic diagnosis of ADPKD. J Am Soc Nephrol 2009; 20:205–212.
- Torres VE, Harris PC, Pirson Y. Autosomal dominant polycystic kidney disease. Lancet 2007; 369:1287–1301.
- Bajwa ZH, Sial KA, Malik AB, Steinman TI. Pain patterns in patients with polycystic kidney disease. Kidney Int 2004; 66:1561–1569.
- Jouret F, Lhommel R, Beguin C, et al. Positron-emission computed tomography in cyst infection diagnosis in patients with autosomal dominant polycystic kidney disease. Clin J Am Soc Nephrol 2011; 6:1644–1650.
- Nishiura JL, Neves RF, Eloi SR, Cintra SM, Ajzen SA, Heilberg IP. Evaluation of nephrolithiasis in autosomal dominant polycystic kidney disease patients. Clin J Am Soc Nephrol 2009; 4:838–844.
- Hiura T, Yamazaki H, Saeki T, et al. Nephrotic syndrome and IgA nephropathy in polycystic kidney disease. Clin Exp Nephrol 2006; 10:136–139.
- Hossack KF, Leddy CL, Johnson AM, Schrier RW, Gabow PA. Echocardiographic findings in autosomal dominant polycystic kidney disease. N Engl J Med 1988; 319:907–912.
- Rossetti S, Chauveau D, Kubly V, et al. Association of mutation position in polycystic kidney disease 1 (PKD1) gene and development of a vascular phenotype. Lancet 2003; 361:2196–2201.
- Linn FH, Wijdicks EF, van der Graaf Y, Weerdesteyn-van Vliet FA, Bartelds AI, van Gijn J. Prospective study of sentinel headache in aneurismal subarachnoid haemorrhage. Lancet 1994; 344:590–593.
- Belz MM, Fick-Brosnahan GM, Hughes RL, et al. Recurrence of intracranial aneurysms in autosomal-dominant polycystic kidney disease. Kidney Int 2003; 63:1824–1830.
- Irazabal MV, Huston J, Kubly V, et al. Extended follow-up of unruptured intracranial aneurysms detected by presymptomatic screening in patients with autosomal dominant polycystic kidney disease. Clin J Am Soc Nephrol 2011; 6:1274–1285.
- Salman A-S, White PM, Counsell CE, et al; Scottish Audit of Intracranial Vascular Malformations Collaborators. Outcome after conservative management or intervention for unruptured brain arteriovenous malformations. JAMA 2014; 311:1661–1669.
- Vijay A, Vijay A, Pankaj P. Autosomal dominant polycystic kidney disease: a comprehensive review. Nephrourol Mon 2010; 2:172–192.
- Grantham JJ, Torres VE, Chapman AB, et al; CRISP Investigators. Volume progression in polycystic kidney disease. N Engl J Med 2006; 354:2122–2130.
- Bae KT, Grantham JJ. Imaging for the prognosis of autosomal dominant polycystic kidney disease. Nat Rev Nephrol 2010; 6:96–106.
- van den Dool SW, Wasser NM, de Fijter JW, Hoekstra J, van der Geest RJ. Functional renal volume: quantitative analysis at gadolinium-enhanced MR angiography—feasibility study in healthy potential kidney donors. Radiology 2005; 236:189–195.
- O’Sullivan DA, Torres VE, Heit JA, Liggett S, King BF. Compression of the inferior vena cava by right renal cysts: an unusual cause of IVC and/or iliofemoral thrombosis with pulmonary embolism in autosomal dominant polycystic kidney disease. Clin Nephrol 1998; 49:332–334.
- Tveit DP, Hypolite I, Bucci J, et al. Risk factors for hospitalizations resulting from pulmonary embolism after renal transplantation in the United States. J Nephrol 2001; 14:361–368.
- Pei Y. A “two-hit” model of cystogenesis in autosomal dominant polycystic kidney disease? Trends Mol Med 2001; 7:151–156.
- Qian F, Germino GG. “Mistakes happen”: somatic mutation and disease. Am J Hum Genet 1997; 61:1000–1005.
- Takakura A, Contrino L, Zhou X, et al. Renal injury is a third hit promoting rapid development of adult polycystic kidney disease. Hum Mol Genet 2009; 18:2523–2531.
- Torres VE, Harris PC. Strategies targeting cAMP signaling in the treatment of polycystic kidney disease. J Am Soc Nephrol 2014; 25:18–32.
- Nauli SM, Alenghat FJ, Luo Y, et al. Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat Genet 2003; 33:129–137.
- Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N Engl J Med 2011; 364:1533–1543.
- Yamaguchi T, Wallace DP, Magenheimer BS, Hempson SJ, Grantham JJ, Calvet JP. Calcium restriction allows cAMP activation of the B-Raf/ERK pathway, switching cells to a cAMP-dependent growth-stimulated phenotype. J Biol Chem 2004; 279:40419–40430.
- Verghese E, Ricardo SD, Weidenfeld R, et al. Renal primary cilia lengthen after acute tubular necrosis. J Am Soc Nephrol 2009; 20:2147–2153.
- Wang X, Wu Y, Ward CJ, Harris PC, Torres VE. Vasopressin directly regulates cyst growth in polycystic kidney disease. J Am Soc Nephrol 2008; 19:102–108.
- Torres VE. Cyclic AMP, at the hub of the cystic cycle. Kidney Int 2004; 66:1283–1285.
- Nagao S, Nishii K, Katsuyama M, et al. Increased water intake decreases progression of polycystic kidney disease in the PCK rat. J Am Soc Nephrol 2006; 17:2220–2227.
- Grantham JJ. Therapy for polycystic kidney disease? It’s water, stupid! J Am Soc Nephrol 2008; 19:1–7.
- Wang CJ, Creed C, Winklhofer FT, Grantham JJ. Water prescription in autosomal dominant polycystic kidney disease: a pilot study. Clin J Am Soc Nephrol 2011; 6:192–197.
- Gattone VH, Wang X, Harris PC, Torres VE. Inhibition of renal cystic disease development and progression by a vasopressin V2 receptor antagonist. Nat Med 2003; 9:1323–1326.
- Torres VE, Chapman AB, Devuyst O, et al; TEMPO 3:4 Trial Investigators. Tolvaptan in patients with autosomal dominant polycystic kidney disease. N Engl J Med 2012; 367:2407–2418.
- Shillingford JM, Murcia NS, Larson CH, et al. The mTOR pathway is regulated by polycystin-1, and its inhibition reverses renal cystogenesis in polycystic kidney disease. Proc Natl Acad Sci U S A 2006; 103:5466–5471.
- Hartman TR, Liu D, Zilfou JT, et al. The tuberous sclerosis proteins regulate formation of the primary cilium via a rapamycin-insensitive and polycystin 1-independent pathway. Hum Mol Genet 2009; 18:161–163.
- Serra AL, Poster D, Kistler AD, et al. Sirolimus and kidney growth in autosomal dominant polycystic kidney disease. N Engl J Med 2010; 363:820–829.
- Walz G, Budde K, Mannaa M, et al. Everolimus in patients with autosomal dominant polycystic kidney disease. N Engl J Med 2010; 363:830–840. Errata in: N Engl J Med 2010; 363:1190 and N Engl J Med 2010; 363:1977.
- Walz G, Budde K, Eckardt K-U. mTOR inhibitors and autosomal dominant polycystic kidney disease (correspondence). N Engl J Med 2011; 364:287–288.
- Braun WE, Schold JD, Stephany BR, Spinko RA, Herfs BR. Low dose rapamycin (sirolimus) effects in autosomal dominant polycystic kidney disease: an open-label randomized control pilot study. Clin J Am Soc Nephrol 2014; 9:881–888.
- Karihaloo A, Koraishy F, Huen SC, et al. Macrophages promote cyst growth in polycystic kidney disease. J Am Soc Nephrol 2011; 22:1809–1814.
- Fox R, Nhan TQ, Law GL, Morris DR, Liles WC, Schwartz SM. PSGL-1 and mTOR regulate translation of ROCK-1 and physiological functions of macrophages. EMBO J 2007; 26:505–515. Erratum in: EMBO J 2007; 26:2605.
- Carvalhosa R, Deambrosis I, Carrera P, et al. Cystogenic potential of CD133+ progenitor cells of human polycystic kidneys. J Pathol 2011; 225:129–141.
- Grantham JJ, Mulamalla S, Grantham CJ, et al. Detected renal cysts are tips of the iceberg in adults with ADPKD. Clin J Am Soc Nephrol 2012; 7:1087–1093.
- Grantham JJ, Cook LT, Wetzel LH, Cadnapaphornchai MA, Bae KT. Evidence of extraordinary growth in the progressive enlargement of renal cysts. Clin J Am Soc Nephrol 2010; 5:889–896.
- James PA, Oparil S, Carter BL, et al. 2014 evidence-based guideline for the management of high blood pressure in adults: report from the panel members appointed to the Eighth Joint National Committee (JNC 8). JAMA 2014; 311:507–520.
- Chapman AB, Torres VE, Perrone RD, et al. The HALT polycystic kidney disease trials: design and implementation. Clin J Am Soc Nephrol 2010; 5:102–109.
- Klahr S, Breyer JA, Beck GJ, et al. Dietary protein restriction, blood pressure control, and the progression of polycystic kidney disease. Modification of Diet in Renal Disease Study Group. J Am Soc Nephrol 1995; 5:2037–2047.
- Thilly N. Low-protein diet in chronic kidney disease: from questions of effectiveness to those of feasibility. Nephrol Dial Transplant 2013; 28:2203–2205.
- Luciano RL, Dahl NK. Extra-renal manifestations of autosomal dominant polycystic kidney disease (ADPKD): considerations for routine screening and management. Nephrol Dial Transplant 2014; 29:247–254.
- Harris PC, Rossetti S. Molecular diagnostics for autosomal dominant polycystic kidney disease. Nat Rev Nephrol 2010; 6:197–206.
- Jacquet A, Pallet N, Kessler M, et al. Outcomes of renal transplantation in patients with autosomal dominant polycystic kidney disease: a nationwide longitudinal study. Transpl Int 2011; 24:582–587.
- Ruggenenti P, Remuzzi A, Ondei P, et al. Safety and efficacy of long-acting somatostatin treatment in autosomal-dominant polycystic kidney disease. Kidney Int 2005; 68:206–216.
- Cadnapaphornchai MA, George DM, McFann K, et al. Effect of pravastatin on total kidney volume, left ventricular mass index, and microalbuminuria in pediatric autosomal dominant polycystic kidney disease. Clin J Am Soc Nephrol 2014; 9:889–896.
- van Dijk MA, Kamper AM, van Veen S, Souverjin JH, Blauw GJ. Effect of simvastatin on renal function in autosomal dominant polycystic kidney disease. Nephrol Dial Transplant 2001; 16:2152–2157.
- Grantham JJ, Uchich M, Cragoe EL, et al. Chemical modification of cell proliferation and fluid secretion in renal cysts. Kidney Int 1989; 35:1379–1389.
- Tanner GA. Potassium citrate/citric acid intake improves renal function in rats with polycystic kidney disease. J Am Soc Nephrol 1998; 9:1242–1248.
- Belibi FA, Edelstein CL. Novel targets for the treatment of autosomal dominant polycystic kidney disease. Expert Opin Investig Drugs 2010; 19:315–328.
- Tao Y, Kim J, Yin Y, et al. VEGF receptor inhibition slows the progression of polycystic kidney disease. Kidney Int 2007; 72:1358–1366.
- Terryn S, Ho A, Beauwens R, Devuyst O. Fluid transport and cystogenesis in autosomal dominant polycystic kidney disease. Biochim Biophys Acta 2011; 1812:1314–1321.
- Thiagarajah JR, Verkman AS. CFTR inhibitors for treating diarrheal disease. Clin Pharmacol Ther 2012; 92:287–290.
- Boehn SN, Spahn S, Neudecker S, et al. Inhibition of Comt with tolcapone slows proression of polycystic kidney disease in the more severely affected PKD/Mhm (cy/+) substrain of the Hannover Sprague-Dawley rat. Nephrol Dial Transplant 2013; 28:2045–2058.
- Rees S, Kittikulsuth W, Roos K, Strait KA, Van Hoek A, Kohan DE. Adenylyl cyclase 6 deficiency ameliorates polycystic kidney disease. J Am Soc Nephrol 2014; 25:232–237.
- Buchholz B, Schley G, Faria D, et al. Hypoxia-inducible factor-1a causes renal cyst expansion through calcium-activated chloride secretion. J Am Soc Nephrol 2014; 25:465–474.
- Wallace DP, White C, Savinkova L, et al. Periostin promotes renal cyst growth and interstitial fibrosis in polycystic kidney disease. Kidney Int 2014; 85:845–854.
- Leuenroth SJ, Crews CM. Targeting cyst initiation in ADPKD. J Am Soc Nephrol 2009; 20:1–3.
KEY POINTS
- For at-risk patients in the previously difficult diagnostic group from 30 to 39 years of age, newer ultrasonographic criteria for diagnosing PKD1 and PKD2 now require a minimum total of three renal cysts.
- An intracranial aneurysm occurs in approximately 16% of ADPKD patients who have a family member with ADPKD plus an intracranial aneurysm or subarachnoid hemorrhage. Appropriate screening is warranted.
- Combined positron-emission and computed tomography helps identify infected renal or liver cysts and may uncover other unsuspected abdominal or pelvic infections.
- Cyst expansion and increasing total kidney volume might be slowed by increasing water intake to 2,500 to 3,000 mL per day, although formal documentation of this is not published. However, this must be done under a physician’s supervision because of possible adverse effects.
- Tolvaptan, a promising new drug for treating ADPKD, failed to receive US approval. Rapamycin is another potentially effective agent but has had mixed results in clinical trials.
Diabetes increases risk of atrial fibrillation
BARCELONA – Adults with diabetes mellitus are at increased risk of subsequent new-onset atrial fibrillation – and the younger the age at diabetes onset, the greater the likelihood of developing the arrhythmia.
That’s the key finding from a Danish national registry study in which all 5,168,416 Danish adults without atrial fibrillation in 1996 were followed through 2012 for development of atrial fibrillation (AF). The study population included 75,197 Danes with diabetes at baseline and another 235,327 who developed the disease during follow-up, Dr. Jannik L. Pallisgaard explained at the annual congress of the European Society of Cardiology.
During follow-up, 5.6% of those with diabetes and 3.3% of those without diabetes developed AF. The mean time from diabetes onset to AF onset was 5 years, reported Dr. Pallisgaard of the University of Copenhagen.
"What was particularly interesting, I think, is that we found the youngest patients were the group at highest risk" of developing AF, he said. "We suggest that starting at the onset of diabetes, routine pulse palpation, ECGs, and focused patient interviews asking about any signs of atrial fibrillation could prove beneficial in detecting the arrhythmia."
The incidence rate ratio for developing AF per 1,000 person-years of follow-up was roughly 2.5-fold greater in 18- to 39-year-olds with diabetes than in their nondiabetic peers. From this peak rate in young adults, the magnitude of relative risk dropped in stepwise fashion with age: The variability in risk was lower in 40- to 60-year-old diabetics than in the 18- to 39-year olds and lower still in 65- to 74-year olds. Variability in the incidence rate ratio finally bottomed out at a still statistically significant 1.3-fold increased risk of developing AF in diabetic individuals ages 75 and older compared to their nondiabetic peers.
Dr. Pallisgaard noted that while the relative risk of developing AF was greatest in the 18- to 39-year-olds, the absolute number of new cases of AF was far greater in older patients because there were so many more of them with diabetes. He cautioned that as the obesity epidemic leads to more and more patients developing type 2 diabetes at younger ages, more cases of AF can be expected in young adults.
Dr. Pallisgaard cited two likely mechanisms underlying the observed increased risk of AF in diabetic patients: left ventricular hypertrophy and vascular inflammation, which are both often present in the diabetic population.
He reported having no financial conflicts regarding this study, conducted with Danish institutional research funds.
BARCELONA – Adults with diabetes mellitus are at increased risk of subsequent new-onset atrial fibrillation – and the younger the age at diabetes onset, the greater the likelihood of developing the arrhythmia.
That’s the key finding from a Danish national registry study in which all 5,168,416 Danish adults without atrial fibrillation in 1996 were followed through 2012 for development of atrial fibrillation (AF). The study population included 75,197 Danes with diabetes at baseline and another 235,327 who developed the disease during follow-up, Dr. Jannik L. Pallisgaard explained at the annual congress of the European Society of Cardiology.
During follow-up, 5.6% of those with diabetes and 3.3% of those without diabetes developed AF. The mean time from diabetes onset to AF onset was 5 years, reported Dr. Pallisgaard of the University of Copenhagen.
"What was particularly interesting, I think, is that we found the youngest patients were the group at highest risk" of developing AF, he said. "We suggest that starting at the onset of diabetes, routine pulse palpation, ECGs, and focused patient interviews asking about any signs of atrial fibrillation could prove beneficial in detecting the arrhythmia."
The incidence rate ratio for developing AF per 1,000 person-years of follow-up was roughly 2.5-fold greater in 18- to 39-year-olds with diabetes than in their nondiabetic peers. From this peak rate in young adults, the magnitude of relative risk dropped in stepwise fashion with age: The variability in risk was lower in 40- to 60-year-old diabetics than in the 18- to 39-year olds and lower still in 65- to 74-year olds. Variability in the incidence rate ratio finally bottomed out at a still statistically significant 1.3-fold increased risk of developing AF in diabetic individuals ages 75 and older compared to their nondiabetic peers.
Dr. Pallisgaard noted that while the relative risk of developing AF was greatest in the 18- to 39-year-olds, the absolute number of new cases of AF was far greater in older patients because there were so many more of them with diabetes. He cautioned that as the obesity epidemic leads to more and more patients developing type 2 diabetes at younger ages, more cases of AF can be expected in young adults.
Dr. Pallisgaard cited two likely mechanisms underlying the observed increased risk of AF in diabetic patients: left ventricular hypertrophy and vascular inflammation, which are both often present in the diabetic population.
He reported having no financial conflicts regarding this study, conducted with Danish institutional research funds.
BARCELONA – Adults with diabetes mellitus are at increased risk of subsequent new-onset atrial fibrillation – and the younger the age at diabetes onset, the greater the likelihood of developing the arrhythmia.
That’s the key finding from a Danish national registry study in which all 5,168,416 Danish adults without atrial fibrillation in 1996 were followed through 2012 for development of atrial fibrillation (AF). The study population included 75,197 Danes with diabetes at baseline and another 235,327 who developed the disease during follow-up, Dr. Jannik L. Pallisgaard explained at the annual congress of the European Society of Cardiology.
During follow-up, 5.6% of those with diabetes and 3.3% of those without diabetes developed AF. The mean time from diabetes onset to AF onset was 5 years, reported Dr. Pallisgaard of the University of Copenhagen.
"What was particularly interesting, I think, is that we found the youngest patients were the group at highest risk" of developing AF, he said. "We suggest that starting at the onset of diabetes, routine pulse palpation, ECGs, and focused patient interviews asking about any signs of atrial fibrillation could prove beneficial in detecting the arrhythmia."
The incidence rate ratio for developing AF per 1,000 person-years of follow-up was roughly 2.5-fold greater in 18- to 39-year-olds with diabetes than in their nondiabetic peers. From this peak rate in young adults, the magnitude of relative risk dropped in stepwise fashion with age: The variability in risk was lower in 40- to 60-year-old diabetics than in the 18- to 39-year olds and lower still in 65- to 74-year olds. Variability in the incidence rate ratio finally bottomed out at a still statistically significant 1.3-fold increased risk of developing AF in diabetic individuals ages 75 and older compared to their nondiabetic peers.
Dr. Pallisgaard noted that while the relative risk of developing AF was greatest in the 18- to 39-year-olds, the absolute number of new cases of AF was far greater in older patients because there were so many more of them with diabetes. He cautioned that as the obesity epidemic leads to more and more patients developing type 2 diabetes at younger ages, more cases of AF can be expected in young adults.
Dr. Pallisgaard cited two likely mechanisms underlying the observed increased risk of AF in diabetic patients: left ventricular hypertrophy and vascular inflammation, which are both often present in the diabetic population.
He reported having no financial conflicts regarding this study, conducted with Danish institutional research funds.
AT THE ESC CONGRESS 2014
Key clinical point: Starting at the onset of diabetes, routine pulse palpation, ECGs, and patient interviews focused on signs of atrial fibrillation might improve detection of the arrhythmia.
Major finding: During follow-up, 5.6% of those with diabetes and 3.3% of those without diabetes developed AF.
Data source: This was a national registry study including all of the nearly 5.2 million Danish adults without atrial fibrillation in 1996. Follow-up ran through 2012.
Disclosures: The presenter reported having no financial conflicts regarding this study, funded by Danish institutional research grants.
Antibody gets orphan status for CTCL in Europe
The European Commission has granted orphan drug designation to IPH4102 for the treatment of cutaneous T-cell lymphoma (CTCL).
IPH4102 is a cytotoxic anti-KIR3DL2 monoclonal antibody (mAb) that targets CTCL cells.
Orphan status provides Innate Pharma, the company developing IPH4102, with benefits such as tax incentives, market exclusivity for 10 years, possibilities for additional research funding, and additional guidance from the European Medicines Agency during clinical development.
Preclinical results with IPH4102 were presented in a poster at the 2014 T-cell Lymphoma Forum. The research was conducted by investigators from Innate Pharma and INSERM at Hôpital Saint Louis in Paris.
The researchers generated 3 mAbs that bind selectively to KIR3DL2 and evaluated their efficacy against KIR3DL2-expressing tumors and Sézary cell lines.
IPH4102 was among the 3 mAbs and emerged as the most promising drug candidate.
Experiments revealed that anti-KIR3DL2 mAbs can kill KIR3DL2+ cell lines through allo-antibody-dependent cell cytotoxicity, even at low tumor antigen density.
The mAbs also improved survival in KIR3DL2+ xenograft models. Survival in mAb-treated mice ranged from 30.5 days to 54.5 days, compared to 19 days in controls.
Finally, the mAbs mediated killing of primary Sézary cells with autologous natural killer cells nearly as efficiently as alemtuzumab.
The investigators said these results suggest anti-KIR3DL2 mAbs are a feasible treatment option for CTCL patients. They plan to prove this hypothesis with a phase 1 trial of IPH4102, which is expected to begin in 2015. ![]()
The European Commission has granted orphan drug designation to IPH4102 for the treatment of cutaneous T-cell lymphoma (CTCL).
IPH4102 is a cytotoxic anti-KIR3DL2 monoclonal antibody (mAb) that targets CTCL cells.
Orphan status provides Innate Pharma, the company developing IPH4102, with benefits such as tax incentives, market exclusivity for 10 years, possibilities for additional research funding, and additional guidance from the European Medicines Agency during clinical development.
Preclinical results with IPH4102 were presented in a poster at the 2014 T-cell Lymphoma Forum. The research was conducted by investigators from Innate Pharma and INSERM at Hôpital Saint Louis in Paris.
The researchers generated 3 mAbs that bind selectively to KIR3DL2 and evaluated their efficacy against KIR3DL2-expressing tumors and Sézary cell lines.
IPH4102 was among the 3 mAbs and emerged as the most promising drug candidate.
Experiments revealed that anti-KIR3DL2 mAbs can kill KIR3DL2+ cell lines through allo-antibody-dependent cell cytotoxicity, even at low tumor antigen density.
The mAbs also improved survival in KIR3DL2+ xenograft models. Survival in mAb-treated mice ranged from 30.5 days to 54.5 days, compared to 19 days in controls.
Finally, the mAbs mediated killing of primary Sézary cells with autologous natural killer cells nearly as efficiently as alemtuzumab.
The investigators said these results suggest anti-KIR3DL2 mAbs are a feasible treatment option for CTCL patients. They plan to prove this hypothesis with a phase 1 trial of IPH4102, which is expected to begin in 2015. ![]()
The European Commission has granted orphan drug designation to IPH4102 for the treatment of cutaneous T-cell lymphoma (CTCL).
IPH4102 is a cytotoxic anti-KIR3DL2 monoclonal antibody (mAb) that targets CTCL cells.
Orphan status provides Innate Pharma, the company developing IPH4102, with benefits such as tax incentives, market exclusivity for 10 years, possibilities for additional research funding, and additional guidance from the European Medicines Agency during clinical development.
Preclinical results with IPH4102 were presented in a poster at the 2014 T-cell Lymphoma Forum. The research was conducted by investigators from Innate Pharma and INSERM at Hôpital Saint Louis in Paris.
The researchers generated 3 mAbs that bind selectively to KIR3DL2 and evaluated their efficacy against KIR3DL2-expressing tumors and Sézary cell lines.
IPH4102 was among the 3 mAbs and emerged as the most promising drug candidate.
Experiments revealed that anti-KIR3DL2 mAbs can kill KIR3DL2+ cell lines through allo-antibody-dependent cell cytotoxicity, even at low tumor antigen density.
The mAbs also improved survival in KIR3DL2+ xenograft models. Survival in mAb-treated mice ranged from 30.5 days to 54.5 days, compared to 19 days in controls.
Finally, the mAbs mediated killing of primary Sézary cells with autologous natural killer cells nearly as efficiently as alemtuzumab.
The investigators said these results suggest anti-KIR3DL2 mAbs are a feasible treatment option for CTCL patients. They plan to prove this hypothesis with a phase 1 trial of IPH4102, which is expected to begin in 2015. ![]()