User login
'Deep learning' AI shows benefit in colonoscopy in U.S. population
Adenoma miss rates were significantly lower with the use of an artificial intelligence (AI)–based computer-aided detection (CADe) system than with high-definition white light (HDWL), according to a new prospective, multicenter, single-blind randomized study based on data from more than 200 colonoscopies.
Missed adenomas can be generally categorized as adenomas fully obscured from the visual field or those appearing partly or fully in the visual field but missed by an endoscopist, wrote Jeremy R. Glissen Brown, MD, of Harvard Medical School, Boston, and colleagues. While retrospective and prospective studies in China, Italy, and Japan have shown that deep-learning CADe improves adenoma identification during colonoscopy, there have been no prospective U.S. studies on CADe in a diverse population, they noted.
In the study published in Clinical Gastroenterology and Hepatology, the researchers reviewed data from 223 adults aged 22 years and older who underwent screening colonoscopies across four U.S. academic medical centers between 2019 and 2020. The procedure indication was primary colorectal cancer screening for 59.6% of the patients and postpolypectomy surveillance for 40.4%. Among this cohort, 45.3% (101) were female, 67.7% (151) were White, and 21% (133) were African American. Participants were randomized to receive either CADe colonoscopy first or HDWL colonoscopy first; the patients immediately underwent the other procedure in tandem fashion from the same endoscopist.
The primary outcome of the study was adenoma miss rate (AMR), defined as “the number of histologically confirmed adenomas detected during the second colonoscopy in either arm divided by the total number of adenomas detected during both procedures.” Sessile serrated lesion (SSL) miss rates and adenomas per colonoscopy (APC) were secondary outcomes.
Overall, the primary outcome of AMR was significantly lower in the CADe-first group, compared with the HDWL-first group (20.12% vs. 31.25%; P = .0247), with an odds ratio of 1.8048 (95% CI, 1.0780-3.0217). The CADe-first group yielded a lower SSL miss rate, compared with the HDLW-first group (7.14% vs. 42.11%; P = .0482), as well as a lower polyp miss rate (20.70% vs. 33.71%; P = .0007). The first-pass number of APC was significantly higher in the CADe-first group, compared with the HDWL-first group (1.19 [SD 2.03] vs. 0.90 [SD 1.55]; P = .0323). In addition, the first-pass adenoma detection rate (ADR) was not significantly different in the CADe-first group, compared with the HDWL-first group (50.44% vs. 43.64%; P = .3091), and the median withdrawal time was significantly shorter with CADe, compared with HDWL (9.5 minutes vs. 8.5 minutes; P = .0098).
There were no significant observable differences between the two groups regarding missed adenomas arranged by size or location. Moreover, there were no significant differences in miss rates for hyperplastic polyps or advanced adenomas. Factors significantly associated with missed adenomas included being in the HDLW-first group, age 65 years or younger, and the right colon vs. other locations. No immediate adverse events occurred in either group.
According to the researchers, while previous studies in China and Italy have shown increased ADR using CADe systems, these results are not generalizable to the U.S. population for several reasons, notably the studies’ inclusion of colonoscopy indications other than colorectal cancer screening and surveillance. Though the present study showed a significantly lower AMR with CADe, it still represents missed adenomas. The researchers note: “In the present study, in which CADe detected 285 polyps, there were only three false negatives (defined as polyps that were visualized by the endoscopist but not by the CADe system). Overall, this suggests that the ‘missed polyps’ in the CADe arm may have been obscured behind folds rather than in the visual field.” They added, “Further research is needed on combining CADe technologies with mucosal exposure devices, as the benefits of these tools for polyp detection may be additive.”
The study findings were limited by several factors, including the inability to detect a difference in overall ADR, the limited generalizability of the tandem study design to real-world practice, the inclusion of only experienced endoscopists, and the use of a second monitor that may have impacted gaze patterns, the researchers noted. However, the results represent the first examination of deep-learning CADe in a diverse U.S. population and showed a decrease in adenoma miss rates and decreased miss rates for polyps and SSLs, compared with HDWL. Based on these findings, the authors concluded CADe “has the potential to decrease inter-provider variability in colonoscopy quality by reducing adenoma miss rate even in experienced providers.”
Reducing miss rates matters
“Missed adenomas can be associated with the development of interval colorectal cancer, so whether novel technologies such as artificial intelligence-based computer-aided polyp detection system can decrease adenoma miss rate is of interest,” said Atsushi Sakuraba, MD, of the University of Chicago, in an interview.
Dr Sakuraba said he was not surprised by the current study findings, as several pilot and randomized studies have shown the benefits of AI-based polyp detection systems. As for how the AI-assisted technology might improve practice, he said it may be a valuable addition. “Adenoma miss rate was significantly lower with an AI-based polyp detection system, so it might lead to decreased colorectal cancer,” he explained. “Various methods to improve adenoma detection should complement each other.
Dr. Sakuraba also commented that additional research is needed outside of academic centers, noting “further studies in the community setting involving various endoscopists are required to confirm generalizability.”
Lead author Dr. Glissen Brown had no financial conflicts to disclose. This was an investigator-initiated study, with research software and study funding provided by Wision. Dr. Sakuraba disclosed collaborative research with Fuji film, which was not involved in this study.
Adenoma miss rates were significantly lower with the use of an artificial intelligence (AI)–based computer-aided detection (CADe) system than with high-definition white light (HDWL), according to a new prospective, multicenter, single-blind randomized study based on data from more than 200 colonoscopies.
Missed adenomas can be generally categorized as adenomas fully obscured from the visual field or those appearing partly or fully in the visual field but missed by an endoscopist, wrote Jeremy R. Glissen Brown, MD, of Harvard Medical School, Boston, and colleagues. While retrospective and prospective studies in China, Italy, and Japan have shown that deep-learning CADe improves adenoma identification during colonoscopy, there have been no prospective U.S. studies on CADe in a diverse population, they noted.
In the study published in Clinical Gastroenterology and Hepatology, the researchers reviewed data from 223 adults aged 22 years and older who underwent screening colonoscopies across four U.S. academic medical centers between 2019 and 2020. The procedure indication was primary colorectal cancer screening for 59.6% of the patients and postpolypectomy surveillance for 40.4%. Among this cohort, 45.3% (101) were female, 67.7% (151) were White, and 21% (133) were African American. Participants were randomized to receive either CADe colonoscopy first or HDWL colonoscopy first; the patients immediately underwent the other procedure in tandem fashion from the same endoscopist.
The primary outcome of the study was adenoma miss rate (AMR), defined as “the number of histologically confirmed adenomas detected during the second colonoscopy in either arm divided by the total number of adenomas detected during both procedures.” Sessile serrated lesion (SSL) miss rates and adenomas per colonoscopy (APC) were secondary outcomes.
Overall, the primary outcome of AMR was significantly lower in the CADe-first group, compared with the HDWL-first group (20.12% vs. 31.25%; P = .0247), with an odds ratio of 1.8048 (95% CI, 1.0780-3.0217). The CADe-first group yielded a lower SSL miss rate, compared with the HDLW-first group (7.14% vs. 42.11%; P = .0482), as well as a lower polyp miss rate (20.70% vs. 33.71%; P = .0007). The first-pass number of APC was significantly higher in the CADe-first group, compared with the HDWL-first group (1.19 [SD 2.03] vs. 0.90 [SD 1.55]; P = .0323). In addition, the first-pass adenoma detection rate (ADR) was not significantly different in the CADe-first group, compared with the HDWL-first group (50.44% vs. 43.64%; P = .3091), and the median withdrawal time was significantly shorter with CADe, compared with HDWL (9.5 minutes vs. 8.5 minutes; P = .0098).
There were no significant observable differences between the two groups regarding missed adenomas arranged by size or location. Moreover, there were no significant differences in miss rates for hyperplastic polyps or advanced adenomas. Factors significantly associated with missed adenomas included being in the HDLW-first group, age 65 years or younger, and the right colon vs. other locations. No immediate adverse events occurred in either group.
According to the researchers, while previous studies in China and Italy have shown increased ADR using CADe systems, these results are not generalizable to the U.S. population for several reasons, notably the studies’ inclusion of colonoscopy indications other than colorectal cancer screening and surveillance. Though the present study showed a significantly lower AMR with CADe, it still represents missed adenomas. The researchers note: “In the present study, in which CADe detected 285 polyps, there were only three false negatives (defined as polyps that were visualized by the endoscopist but not by the CADe system). Overall, this suggests that the ‘missed polyps’ in the CADe arm may have been obscured behind folds rather than in the visual field.” They added, “Further research is needed on combining CADe technologies with mucosal exposure devices, as the benefits of these tools for polyp detection may be additive.”
The study findings were limited by several factors, including the inability to detect a difference in overall ADR, the limited generalizability of the tandem study design to real-world practice, the inclusion of only experienced endoscopists, and the use of a second monitor that may have impacted gaze patterns, the researchers noted. However, the results represent the first examination of deep-learning CADe in a diverse U.S. population and showed a decrease in adenoma miss rates and decreased miss rates for polyps and SSLs, compared with HDWL. Based on these findings, the authors concluded CADe “has the potential to decrease inter-provider variability in colonoscopy quality by reducing adenoma miss rate even in experienced providers.”
Reducing miss rates matters
“Missed adenomas can be associated with the development of interval colorectal cancer, so whether novel technologies such as artificial intelligence-based computer-aided polyp detection system can decrease adenoma miss rate is of interest,” said Atsushi Sakuraba, MD, of the University of Chicago, in an interview.
Dr Sakuraba said he was not surprised by the current study findings, as several pilot and randomized studies have shown the benefits of AI-based polyp detection systems. As for how the AI-assisted technology might improve practice, he said it may be a valuable addition. “Adenoma miss rate was significantly lower with an AI-based polyp detection system, so it might lead to decreased colorectal cancer,” he explained. “Various methods to improve adenoma detection should complement each other.
Dr. Sakuraba also commented that additional research is needed outside of academic centers, noting “further studies in the community setting involving various endoscopists are required to confirm generalizability.”
Lead author Dr. Glissen Brown had no financial conflicts to disclose. This was an investigator-initiated study, with research software and study funding provided by Wision. Dr. Sakuraba disclosed collaborative research with Fuji film, which was not involved in this study.
Adenoma miss rates were significantly lower with the use of an artificial intelligence (AI)–based computer-aided detection (CADe) system than with high-definition white light (HDWL), according to a new prospective, multicenter, single-blind randomized study based on data from more than 200 colonoscopies.
Missed adenomas can be generally categorized as adenomas fully obscured from the visual field or those appearing partly or fully in the visual field but missed by an endoscopist, wrote Jeremy R. Glissen Brown, MD, of Harvard Medical School, Boston, and colleagues. While retrospective and prospective studies in China, Italy, and Japan have shown that deep-learning CADe improves adenoma identification during colonoscopy, there have been no prospective U.S. studies on CADe in a diverse population, they noted.
In the study published in Clinical Gastroenterology and Hepatology, the researchers reviewed data from 223 adults aged 22 years and older who underwent screening colonoscopies across four U.S. academic medical centers between 2019 and 2020. The procedure indication was primary colorectal cancer screening for 59.6% of the patients and postpolypectomy surveillance for 40.4%. Among this cohort, 45.3% (101) were female, 67.7% (151) were White, and 21% (133) were African American. Participants were randomized to receive either CADe colonoscopy first or HDWL colonoscopy first; the patients immediately underwent the other procedure in tandem fashion from the same endoscopist.
The primary outcome of the study was adenoma miss rate (AMR), defined as “the number of histologically confirmed adenomas detected during the second colonoscopy in either arm divided by the total number of adenomas detected during both procedures.” Sessile serrated lesion (SSL) miss rates and adenomas per colonoscopy (APC) were secondary outcomes.
Overall, the primary outcome of AMR was significantly lower in the CADe-first group, compared with the HDWL-first group (20.12% vs. 31.25%; P = .0247), with an odds ratio of 1.8048 (95% CI, 1.0780-3.0217). The CADe-first group yielded a lower SSL miss rate, compared with the HDLW-first group (7.14% vs. 42.11%; P = .0482), as well as a lower polyp miss rate (20.70% vs. 33.71%; P = .0007). The first-pass number of APC was significantly higher in the CADe-first group, compared with the HDWL-first group (1.19 [SD 2.03] vs. 0.90 [SD 1.55]; P = .0323). In addition, the first-pass adenoma detection rate (ADR) was not significantly different in the CADe-first group, compared with the HDWL-first group (50.44% vs. 43.64%; P = .3091), and the median withdrawal time was significantly shorter with CADe, compared with HDWL (9.5 minutes vs. 8.5 minutes; P = .0098).
There were no significant observable differences between the two groups regarding missed adenomas arranged by size or location. Moreover, there were no significant differences in miss rates for hyperplastic polyps or advanced adenomas. Factors significantly associated with missed adenomas included being in the HDLW-first group, age 65 years or younger, and the right colon vs. other locations. No immediate adverse events occurred in either group.
According to the researchers, while previous studies in China and Italy have shown increased ADR using CADe systems, these results are not generalizable to the U.S. population for several reasons, notably the studies’ inclusion of colonoscopy indications other than colorectal cancer screening and surveillance. Though the present study showed a significantly lower AMR with CADe, it still represents missed adenomas. The researchers note: “In the present study, in which CADe detected 285 polyps, there were only three false negatives (defined as polyps that were visualized by the endoscopist but not by the CADe system). Overall, this suggests that the ‘missed polyps’ in the CADe arm may have been obscured behind folds rather than in the visual field.” They added, “Further research is needed on combining CADe technologies with mucosal exposure devices, as the benefits of these tools for polyp detection may be additive.”
The study findings were limited by several factors, including the inability to detect a difference in overall ADR, the limited generalizability of the tandem study design to real-world practice, the inclusion of only experienced endoscopists, and the use of a second monitor that may have impacted gaze patterns, the researchers noted. However, the results represent the first examination of deep-learning CADe in a diverse U.S. population and showed a decrease in adenoma miss rates and decreased miss rates for polyps and SSLs, compared with HDWL. Based on these findings, the authors concluded CADe “has the potential to decrease inter-provider variability in colonoscopy quality by reducing adenoma miss rate even in experienced providers.”
Reducing miss rates matters
“Missed adenomas can be associated with the development of interval colorectal cancer, so whether novel technologies such as artificial intelligence-based computer-aided polyp detection system can decrease adenoma miss rate is of interest,” said Atsushi Sakuraba, MD, of the University of Chicago, in an interview.
Dr Sakuraba said he was not surprised by the current study findings, as several pilot and randomized studies have shown the benefits of AI-based polyp detection systems. As for how the AI-assisted technology might improve practice, he said it may be a valuable addition. “Adenoma miss rate was significantly lower with an AI-based polyp detection system, so it might lead to decreased colorectal cancer,” he explained. “Various methods to improve adenoma detection should complement each other.
Dr. Sakuraba also commented that additional research is needed outside of academic centers, noting “further studies in the community setting involving various endoscopists are required to confirm generalizability.”
Lead author Dr. Glissen Brown had no financial conflicts to disclose. This was an investigator-initiated study, with research software and study funding provided by Wision. Dr. Sakuraba disclosed collaborative research with Fuji film, which was not involved in this study.
FROM CLINICAL GASTROENTEROLOGY AND HEPATOLOGY
Concordance of DNA Repair Gene Mutations in Paired Primary Prostate Cancer Samples and Metastatic Tissue or Cell-free DNA
Importance
DNA damage response repair (DDR) gene mutations represent actionable alterations that can guide precision medicine strategies in men with advanced prostate cancer (PC). However, acquisition of contemporary tissue samples for molecular testing can be a barrier to deploying precision medicine approaches. We hypothesized that DDR alterations represent truncal events in PC and that primary tissue would reflect mutations found in cell-free circulating tumor (ctDNA) and/or metastatic tissue. OBJECTIVE: To assess concordance in DDR gene alterations between primary PC and metastases or ctDNA specimens.
Methods
Patients were included if a DDR pathway mutation was detected in metastatic tissue or ctDNA and primary tissue sequencing was available for comparison. Sequencing data from three cohorts were analyzed: (1) FoundationOne; (2) University of Washington (UW-OncoPlex or SU2C/PCF International Dream Team sequencing pipelines); and (3) University of Washington rapid autopsy series. Only pathogenic somatic mutations were included and we required 30 days between primary tumor tissue and ctDNA/ metastatic tissue acquisition. Clonal hematopoiesis of indeterminant potential (CHIP) and germline events were adjudicated by an expert molecular pathologist and excluded. DDR gene mutations detected in primary prostate tissue matched with metastatic tissue and/or ctDNA findings.
Results
Paired primary and ctDNA/metastatic samples were sequenced from 72 individuals with known DDR alterations. After excluding ctDNA studies where only CHIP and/or germline events (N=21) were observed, 51 subjects remained and were included in the final analysis. The median time from acquisition of primary tissue to acquisition of ctDNA or tumor tissue was 55 months (range: 5-193 months). Concordance in DDR gene mutation status across samples was 84% (95% CI: 71-92%). Rates of concordance between metastatic-primary and ctDNAprimary pairs were similar when CHIP cases were excluded. BRCA2 reversion mutations associated with resistance to PARP inhibitors and platinum chemotherapy were detected in ctDNA from two subjects.
Discussion
Primary prostate tissue accurately reflected the mutational status of actionable DDR genes in metastatic tissue, consistent with DDR alterations being truncal in most cases. After excluding likely CHIP events, ctDNA profiling accurately captured these DDR mutations, while also detecting reversion alterations that may suggest resistance mechanisms.
Importance
DNA damage response repair (DDR) gene mutations represent actionable alterations that can guide precision medicine strategies in men with advanced prostate cancer (PC). However, acquisition of contemporary tissue samples for molecular testing can be a barrier to deploying precision medicine approaches. We hypothesized that DDR alterations represent truncal events in PC and that primary tissue would reflect mutations found in cell-free circulating tumor (ctDNA) and/or metastatic tissue. OBJECTIVE: To assess concordance in DDR gene alterations between primary PC and metastases or ctDNA specimens.
Methods
Patients were included if a DDR pathway mutation was detected in metastatic tissue or ctDNA and primary tissue sequencing was available for comparison. Sequencing data from three cohorts were analyzed: (1) FoundationOne; (2) University of Washington (UW-OncoPlex or SU2C/PCF International Dream Team sequencing pipelines); and (3) University of Washington rapid autopsy series. Only pathogenic somatic mutations were included and we required 30 days between primary tumor tissue and ctDNA/ metastatic tissue acquisition. Clonal hematopoiesis of indeterminant potential (CHIP) and germline events were adjudicated by an expert molecular pathologist and excluded. DDR gene mutations detected in primary prostate tissue matched with metastatic tissue and/or ctDNA findings.
Results
Paired primary and ctDNA/metastatic samples were sequenced from 72 individuals with known DDR alterations. After excluding ctDNA studies where only CHIP and/or germline events (N=21) were observed, 51 subjects remained and were included in the final analysis. The median time from acquisition of primary tissue to acquisition of ctDNA or tumor tissue was 55 months (range: 5-193 months). Concordance in DDR gene mutation status across samples was 84% (95% CI: 71-92%). Rates of concordance between metastatic-primary and ctDNAprimary pairs were similar when CHIP cases were excluded. BRCA2 reversion mutations associated with resistance to PARP inhibitors and platinum chemotherapy were detected in ctDNA from two subjects.
Discussion
Primary prostate tissue accurately reflected the mutational status of actionable DDR genes in metastatic tissue, consistent with DDR alterations being truncal in most cases. After excluding likely CHIP events, ctDNA profiling accurately captured these DDR mutations, while also detecting reversion alterations that may suggest resistance mechanisms.
Importance
DNA damage response repair (DDR) gene mutations represent actionable alterations that can guide precision medicine strategies in men with advanced prostate cancer (PC). However, acquisition of contemporary tissue samples for molecular testing can be a barrier to deploying precision medicine approaches. We hypothesized that DDR alterations represent truncal events in PC and that primary tissue would reflect mutations found in cell-free circulating tumor (ctDNA) and/or metastatic tissue. OBJECTIVE: To assess concordance in DDR gene alterations between primary PC and metastases or ctDNA specimens.
Methods
Patients were included if a DDR pathway mutation was detected in metastatic tissue or ctDNA and primary tissue sequencing was available for comparison. Sequencing data from three cohorts were analyzed: (1) FoundationOne; (2) University of Washington (UW-OncoPlex or SU2C/PCF International Dream Team sequencing pipelines); and (3) University of Washington rapid autopsy series. Only pathogenic somatic mutations were included and we required 30 days between primary tumor tissue and ctDNA/ metastatic tissue acquisition. Clonal hematopoiesis of indeterminant potential (CHIP) and germline events were adjudicated by an expert molecular pathologist and excluded. DDR gene mutations detected in primary prostate tissue matched with metastatic tissue and/or ctDNA findings.
Results
Paired primary and ctDNA/metastatic samples were sequenced from 72 individuals with known DDR alterations. After excluding ctDNA studies where only CHIP and/or germline events (N=21) were observed, 51 subjects remained and were included in the final analysis. The median time from acquisition of primary tissue to acquisition of ctDNA or tumor tissue was 55 months (range: 5-193 months). Concordance in DDR gene mutation status across samples was 84% (95% CI: 71-92%). Rates of concordance between metastatic-primary and ctDNAprimary pairs were similar when CHIP cases were excluded. BRCA2 reversion mutations associated with resistance to PARP inhibitors and platinum chemotherapy were detected in ctDNA from two subjects.
Discussion
Primary prostate tissue accurately reflected the mutational status of actionable DDR genes in metastatic tissue, consistent with DDR alterations being truncal in most cases. After excluding likely CHIP events, ctDNA profiling accurately captured these DDR mutations, while also detecting reversion alterations that may suggest resistance mechanisms.
Methods of Identifying Real World mCRPC Patients from the Veterans Health Administration System
Purpose
Prostate cancer is the fifth leading cause of death in the United States. Genomic testing is essential to guide treatment decisions in patients with metastatic castration resistant prostate cancer (mCRPC), the most advanced stage of prostate cancer. However, identifying mCRPC patients from administrative data is challenging and hinders researchers’ ability to assess testing among these patients. This study aims to develop algorithms using structured data and unstructured data with Natural language processing (NLP) methods to identify veterans by disease stage and hormone sensitivity, and to assess patient characteristics as well as receipt of tumor NGS testing.
Methods
We used biopsy, pathology, and diagnosis codes, to identify veterans with newly diagnosed PC within the Veterans Health Administration (VA) from January 1, 2017 to December 31, 2020. We developed and deployed: 1. A structured algorithm that used medication and Prostate-Specific Antigen (PSA) data to assess hormone sensitivity. 2. NLP tools to extract disease stage and hormone sensitivity from clinical notes. We report descriptive statistics on patient demographics, clinical characteristics, disease status, androgen deprivation therapy (ADT), and receipt of tumor NGS testing.
Results
There were 42,485 veterans with newly diagnosed prostate cancer between 2017-2020. This represented ~0.18% of veterans served in the VA and consisted of Whites (57%), Blacks (33%), and others (10%). During the study period, 3,113 (7.3%) patients had documentation of assessment for intraductal carcinoma, 5,160 (12.1%) had ADT treatment, 1,481 (3.5%) had CRPC, and 3,246 (7.6%) had metastatic disease. Among the 42,485 veterans, 422 received tumor NGS testing within VA, and 300 of them had metastatic disease. NLP tool and structured data algorithm collectively showed that 38% of the 422 tumor NGS testing recipients had mCRPC. Among all newly diagnosed PC patients, White patients had highest rates of tumor-based testing (2.3%), then Native Hawaiians (1.7%), Asians and Blacks (1.2% each), compared to Native Americans (0.4%).
Implications
NLP tools alongside structured data algorithms successfully identified variables required to measure access to tumor NGS testing. Efforts to validate and apply this method is ongoing to assess receipt of precision prostate cancer care in VA.
Purpose
Prostate cancer is the fifth leading cause of death in the United States. Genomic testing is essential to guide treatment decisions in patients with metastatic castration resistant prostate cancer (mCRPC), the most advanced stage of prostate cancer. However, identifying mCRPC patients from administrative data is challenging and hinders researchers’ ability to assess testing among these patients. This study aims to develop algorithms using structured data and unstructured data with Natural language processing (NLP) methods to identify veterans by disease stage and hormone sensitivity, and to assess patient characteristics as well as receipt of tumor NGS testing.
Methods
We used biopsy, pathology, and diagnosis codes, to identify veterans with newly diagnosed PC within the Veterans Health Administration (VA) from January 1, 2017 to December 31, 2020. We developed and deployed: 1. A structured algorithm that used medication and Prostate-Specific Antigen (PSA) data to assess hormone sensitivity. 2. NLP tools to extract disease stage and hormone sensitivity from clinical notes. We report descriptive statistics on patient demographics, clinical characteristics, disease status, androgen deprivation therapy (ADT), and receipt of tumor NGS testing.
Results
There were 42,485 veterans with newly diagnosed prostate cancer between 2017-2020. This represented ~0.18% of veterans served in the VA and consisted of Whites (57%), Blacks (33%), and others (10%). During the study period, 3,113 (7.3%) patients had documentation of assessment for intraductal carcinoma, 5,160 (12.1%) had ADT treatment, 1,481 (3.5%) had CRPC, and 3,246 (7.6%) had metastatic disease. Among the 42,485 veterans, 422 received tumor NGS testing within VA, and 300 of them had metastatic disease. NLP tool and structured data algorithm collectively showed that 38% of the 422 tumor NGS testing recipients had mCRPC. Among all newly diagnosed PC patients, White patients had highest rates of tumor-based testing (2.3%), then Native Hawaiians (1.7%), Asians and Blacks (1.2% each), compared to Native Americans (0.4%).
Implications
NLP tools alongside structured data algorithms successfully identified variables required to measure access to tumor NGS testing. Efforts to validate and apply this method is ongoing to assess receipt of precision prostate cancer care in VA.
Purpose
Prostate cancer is the fifth leading cause of death in the United States. Genomic testing is essential to guide treatment decisions in patients with metastatic castration resistant prostate cancer (mCRPC), the most advanced stage of prostate cancer. However, identifying mCRPC patients from administrative data is challenging and hinders researchers’ ability to assess testing among these patients. This study aims to develop algorithms using structured data and unstructured data with Natural language processing (NLP) methods to identify veterans by disease stage and hormone sensitivity, and to assess patient characteristics as well as receipt of tumor NGS testing.
Methods
We used biopsy, pathology, and diagnosis codes, to identify veterans with newly diagnosed PC within the Veterans Health Administration (VA) from January 1, 2017 to December 31, 2020. We developed and deployed: 1. A structured algorithm that used medication and Prostate-Specific Antigen (PSA) data to assess hormone sensitivity. 2. NLP tools to extract disease stage and hormone sensitivity from clinical notes. We report descriptive statistics on patient demographics, clinical characteristics, disease status, androgen deprivation therapy (ADT), and receipt of tumor NGS testing.
Results
There were 42,485 veterans with newly diagnosed prostate cancer between 2017-2020. This represented ~0.18% of veterans served in the VA and consisted of Whites (57%), Blacks (33%), and others (10%). During the study period, 3,113 (7.3%) patients had documentation of assessment for intraductal carcinoma, 5,160 (12.1%) had ADT treatment, 1,481 (3.5%) had CRPC, and 3,246 (7.6%) had metastatic disease. Among the 42,485 veterans, 422 received tumor NGS testing within VA, and 300 of them had metastatic disease. NLP tool and structured data algorithm collectively showed that 38% of the 422 tumor NGS testing recipients had mCRPC. Among all newly diagnosed PC patients, White patients had highest rates of tumor-based testing (2.3%), then Native Hawaiians (1.7%), Asians and Blacks (1.2% each), compared to Native Americans (0.4%).
Implications
NLP tools alongside structured data algorithms successfully identified variables required to measure access to tumor NGS testing. Efforts to validate and apply this method is ongoing to assess receipt of precision prostate cancer care in VA.
Diagnosis of Prostate Cancer and Prostate-specific Antigen Level on Initial Prostate Biopsy: Does Race Matter?
Objective
To determine whether Black Veterans are at higher risk for prostate cancer diagnosis on their first prostate biopsy compared to non-Hispanic White (White) Veterans.
Background
Prostate-specific antigen (PSA) testing is widely used to screen for prostate cancer. Although men of African ancestry display an increased incidence of prostate cancer and more aggressive disease, specific PSA thresholds for biopsy referral have yet to be proposed for this population.
Methods
We used the VHA’s electronic medical record data to collect Veterans’ demographic and clinical characteristics including self-identified race/ethnicity, age, date of first prostate biopsy, PSA results, and prostate cancer diagnosis. Veterans’ ZIP code of residence was used to determine urban/rural status, income, and education. We estimated multivariable logistic regression models to predict the likelihood of prostate cancer diagnosis on the first biopsy using race, baseline PSA, age at first PSA test, age at initial biopsy, smoking status, use of statins, and socioeconomic factors as predictors. We calculated adjusted predicted probabilities of cancer detection on the first prostate biopsy from the logistic models at different PSA levels.
Results
We identified 246,056 White and 71,653 Black Veterans who underwent their first prostate biopsy through February 28, 2020 and who had no previous prostate cancer diagnosis or treatment prior to that biopsy. Black Veterans appeared to receive their first PSA test four years earlier and undergo their first prostate biopsy two years earlier than their White counterparts (median age of 57 vs. 61 and 63 vs. 65, respectively). After controlling for selected covariates, we found that Black Veterans were 52% more likely to be diagnosed with prostate cancer on their first prostate biopsy compared to White Veterans (OR 1.52, 95% CI 1.49-1.55). Our model indicated that a Black Veteran with a PSA of 4.0 ng/ml has an equivalent risk of prostate cancer detection as a White Veteran with a PSA of 9.7 ng/ml.
Implications
Our findings suggested that developing a risk-based PSA threshold for referral to prostate biopsy may lead to earlier diagnosis of clinically significant prostate cancer in a population of Veterans known to have an increased incidence and risk of aggressive disease.
Objective
To determine whether Black Veterans are at higher risk for prostate cancer diagnosis on their first prostate biopsy compared to non-Hispanic White (White) Veterans.
Background
Prostate-specific antigen (PSA) testing is widely used to screen for prostate cancer. Although men of African ancestry display an increased incidence of prostate cancer and more aggressive disease, specific PSA thresholds for biopsy referral have yet to be proposed for this population.
Methods
We used the VHA’s electronic medical record data to collect Veterans’ demographic and clinical characteristics including self-identified race/ethnicity, age, date of first prostate biopsy, PSA results, and prostate cancer diagnosis. Veterans’ ZIP code of residence was used to determine urban/rural status, income, and education. We estimated multivariable logistic regression models to predict the likelihood of prostate cancer diagnosis on the first biopsy using race, baseline PSA, age at first PSA test, age at initial biopsy, smoking status, use of statins, and socioeconomic factors as predictors. We calculated adjusted predicted probabilities of cancer detection on the first prostate biopsy from the logistic models at different PSA levels.
Results
We identified 246,056 White and 71,653 Black Veterans who underwent their first prostate biopsy through February 28, 2020 and who had no previous prostate cancer diagnosis or treatment prior to that biopsy. Black Veterans appeared to receive their first PSA test four years earlier and undergo their first prostate biopsy two years earlier than their White counterparts (median age of 57 vs. 61 and 63 vs. 65, respectively). After controlling for selected covariates, we found that Black Veterans were 52% more likely to be diagnosed with prostate cancer on their first prostate biopsy compared to White Veterans (OR 1.52, 95% CI 1.49-1.55). Our model indicated that a Black Veteran with a PSA of 4.0 ng/ml has an equivalent risk of prostate cancer detection as a White Veteran with a PSA of 9.7 ng/ml.
Implications
Our findings suggested that developing a risk-based PSA threshold for referral to prostate biopsy may lead to earlier diagnosis of clinically significant prostate cancer in a population of Veterans known to have an increased incidence and risk of aggressive disease.
Objective
To determine whether Black Veterans are at higher risk for prostate cancer diagnosis on their first prostate biopsy compared to non-Hispanic White (White) Veterans.
Background
Prostate-specific antigen (PSA) testing is widely used to screen for prostate cancer. Although men of African ancestry display an increased incidence of prostate cancer and more aggressive disease, specific PSA thresholds for biopsy referral have yet to be proposed for this population.
Methods
We used the VHA’s electronic medical record data to collect Veterans’ demographic and clinical characteristics including self-identified race/ethnicity, age, date of first prostate biopsy, PSA results, and prostate cancer diagnosis. Veterans’ ZIP code of residence was used to determine urban/rural status, income, and education. We estimated multivariable logistic regression models to predict the likelihood of prostate cancer diagnosis on the first biopsy using race, baseline PSA, age at first PSA test, age at initial biopsy, smoking status, use of statins, and socioeconomic factors as predictors. We calculated adjusted predicted probabilities of cancer detection on the first prostate biopsy from the logistic models at different PSA levels.
Results
We identified 246,056 White and 71,653 Black Veterans who underwent their first prostate biopsy through February 28, 2020 and who had no previous prostate cancer diagnosis or treatment prior to that biopsy. Black Veterans appeared to receive their first PSA test four years earlier and undergo their first prostate biopsy two years earlier than their White counterparts (median age of 57 vs. 61 and 63 vs. 65, respectively). After controlling for selected covariates, we found that Black Veterans were 52% more likely to be diagnosed with prostate cancer on their first prostate biopsy compared to White Veterans (OR 1.52, 95% CI 1.49-1.55). Our model indicated that a Black Veteran with a PSA of 4.0 ng/ml has an equivalent risk of prostate cancer detection as a White Veteran with a PSA of 9.7 ng/ml.
Implications
Our findings suggested that developing a risk-based PSA threshold for referral to prostate biopsy may lead to earlier diagnosis of clinically significant prostate cancer in a population of Veterans known to have an increased incidence and risk of aggressive disease.
Bone Health in Patients With Prostate Cancer: An Evidence-Based Algorithm
Prostate cancer (PC) is the most commonly and newly diagnosed nonskin cancer and the second leading cause of cancer death in men in the United States. About 191,930 cases and about 33,330 deaths from PC were expected for the year 2020.1 About 1 in 41 men will die of PC. Most men diagnosed with PC are aged > 65 years and do not die of their disease. The 5-year survival rate of localized and regional disease is nearly 100%, and disease with distant metastases is 31%. As a result, more than 3.1 million men in the United States who have been diagnosed with PC are still alive today.1 Among veterans, there is a substantial population living with PC. Skolarus and Hawley reported in 2014 that an estimated 200,000 veterans with PC were survivors and 12,000 were newly diagnosed.2
In PC, skeletal strength can be affected by several factors, such as aging, malnutrition, androgen-deprivation therapy (ADT), and bone metastasis.3,4 In fact, most men can live the rest of their life with PC by using strategies to monitor and treat it, once it shows either radiographic or chemical signs of progression.5 ADT is the standard of care to treat hormone-sensitive PC, which is associated with significant skeletal-related adverse effects (AEs).6,7
Men undergoing ADT are 4 times more likely to develop substantial bone deficiency, Shahinian and colleagues found that in men surviving 5 years after PC diagnosis, 19.4% of those who received ADT had a fracture compared with 12% in men who did not (P < .001). The authors established a significant relation between the number of doses of gonadotropin-releasing hormone given in the first 12 months and the risk of fracture.8 Of those who progressed to metastatic disease, the first metastatic nonnodal site is most commonly to the bone.9 Advanced PC is characterized by increased bone turnover, which further raises concerns for bone health and patient performance.10
Skeletal-related events (SREs) include pathologic fracture, spinal cord compression, palliative radiation, or surgery to bone, and change in antineoplastic therapy secondary to bone pain. The concept of bone health refers to the prevention, diagnosis, and treatment of idiopathic, pathogenic, and treatment-related bone loss and delay or prevention of SREs.6,11 Guidelines and expert groups have recommended screening for osteoporosis at the start of ADT with bone mineral density testing, ensuring adequate calcium and vitamin D intake, modifying lifestyle behaviors (smoking cessation, alcohol moderation, and regular exercise), and prescribing bisphosphonates or receptor-activated nuclear factor κ-B ligand inhibitor, denosumab, for men with osteoporosis or who are at general high-fracture risk.12,13 The overuse of these medications results in undue cost to patients as well as AEs, such as osteonecrosis of the jaw (ONJ), hypocalcemia, and bone/joint pains.14-17 There are evidence-based guidelines for appropriate use of bisphosphonates and denosumab for delay and prevention of SREs in the setting of advanced PC.18 These doses also typically differ in frequency to those of osteoporosis.19 We summarize the evidence and guidance for health care providers who care for patients with PC at various stages and complications from both disease-related and treatment-related comorbidities.
Bone-Strengthening Agents
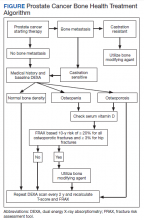
Overall, there is evidence to support the use of bone-strengthening agents in patients with osteopenia/osteoporosis in the prevention of SREs with significant risk factors for progressive bone demineralization, such as lifestyle factors and, in particular, treatments such as ADT. Bone-remodeling agents for treatment of bony metastasis have been shown to provide therapeutic advantage only in limited instances in the castration-resistant PC (CRPC) setting. Hence, in patients with hormone-sensitive PC due to medication-related AEs, treatment with bone-strengthening agents is indicated only if the patient has a significant preexisting risk for fracture from osteopenia/osteoporosis (Table). The Figure depicts an algorithm for the management of bone health in men with PC who are being treated with ADT.
Denosumab and bisphosphonates have an established role in preventing SREs in metastatic CRPC.20 The choice of denosumab or a bisphosphonate typically varies based on the indication, possible AEs, and cost of therapy. There are multiple studies involving initiation of these agents at various stages of disease to improve both time to progression as well as management of SREs. There is a lack of evidence that bisphosphonates prevent metastatic-bone lesions in castration-sensitive PC; therefore, prophylactic use of this agent is not recommended in patients unless they have significant bone demineralization.21,22
Medication-induced ONJ is a severe AE of both denosumab and bisphosphonate therapies. Data from recent trials showed that higher dosing and prolonged duration of denosumab and bisphosphonate therapies further increased risk of ONJ by 1.8% and 1.3%, respectively.15 Careful history taking and discussions with the patient and if possible their dentist on how to reduce risk are recommended. It is good practice for the patient to complete a dental evaluation prior to starting IV bisphosphonates or denosumab. Dental evaluations should be performed routinely at 3- to 12-month intervals throughout therapy based on individualized risk assessment.23 The benefits of using bisphosphonates to prevent fractures associated with osteoporosis outweigh the risk of ONJ in high-risk populations, but not in all patients with PC. A case-by-case basis and evaluation of risk factors should be performed prior to administering bone-modifying therapy. The long-term safety of IV bisphosphonates has not been adequately studied in controlled trials, and concerns regarding long-term complications, including renal toxicity, ONJ, and atypical femoral fractures, remain with prolonged therapy.24,25
The CALGB 70604 (Alliance) trial compared 3-month dosing to monthly treatment with zoledronic acid (ZA), showing no inferiority to lower frequency dosing.26 A Cochrane review of clinical trials found that in patients with advanced PC, bisphosphonates were found to provide roughly 58 fewer SREs per 1000 on average.27 A phase 3 study showed a modest benefit to denosumab vs ZA in the CRPC group regarding incidence of SREs. The rates of SREs were 289 of 951 patients in the bisphosphonate group, and 241 of 950 patients in the denosumab group (30.4% vs 25.3%; hazard ratio [HR], 0.78; 95% CI, 0.66-0.93; P = .005).28 In 2020, the American Society of Clinical Oncology endorsed the Cancer Care Ontario guidelines for prostate bone health care.18 Adequate supplementation is necessary in all patients treated with a bisphosphonate or denosumab to prevent treatment-related hypocalcemia. Typically, daily supplementation with a minimum of calcium 500 mg and vitamin D 400 IU is recommended.16
Bone Health in Patients
Nonmetastatic Hormone-Sensitive PC
ADT forms the backbone of treatment for patients with local and advanced metastatic castration-sensitive PC along with surgical and focal radiotherapy options. Cancer treatment-induced bone loss is known to occur with prolonged use of ADT. The ZEUS trial found no prevention of bone metastasis in patients with high-risk localized PC with the use of ZA in the absence of bone metastasis. A Kaplan-Meier estimated proportion of bone metastases after a median follow-up of 4.8 years was found to be not statistically significant: 14.7% in the ZA group vs 13.2% in the control/placebo group.29 The STAMPEDE trial showed no significant overall survival (OS) benefit with the addition of ZA to ADT vs ADT alone (HR, 0.94; 95% CI, 0.79-1.11; P = .45), 5-year survival with ADT alone was 55% compared to ADT plus ZA with 57% 5-year survival.30 The RADAR trial showed that at 5 years in high Gleason score patients, use of ZA in the absence of bone metastasis was beneficial, but not in low- or intermediate-risk patients. However, at 10-year analysis there was no significant difference in any of the high-stratified groups with or without ZA.31
The PR04 trial showed no effect on OS with clodronate compared with placebo in nonmetastatic castration-sensitive PC, with a HR of 1.12 (95% CI, 0.89-1.42; P = .94). The estimated 5-year survival was 80% with placebo and 78% with clodronate; 10-year survival rates were 51% with placebo and 48% with clodronate.32 Data from the HALT trial showed an increased bone mineral density and reduced risk of new vertebral fractures vs placebo (1.5% vs 3.9%, respectively) in the absence of metastatic bone lesions and a reduction in new vertebral fractures in patients with nonmetastatic PC.33 Most of these studies showed no benefit with the addition of ZA to nonmetastatic PC; although, the HALT trial provides evidence to support use of denosumab in patients with nonmetastatic PC for preventing vertebral fragility fractures in men receiving ADT.
Metastatic Hormone-Sensitive PC
ZA is often used to treat men with metastatic castration-sensitive PC despite limited efficacy and safety data. The CALGB 90202 (Alliance) trial authors found that the early use of ZA was not associated with increased time to first SRE. The median time to first SRE was 31.9 months in the ZA group (95% CI, 24.2-40.3) and 29.8 months in the placebo group (stratified HR, 0.97; 95% CI, 0-1.17; 1-sided stratified log-rank P = .39).34 OS was similar between the groups (HR, 0.88; 95% CI, 0.70-1.12; P = .29) as were reported AEs.34 Results from these studies suggest limited benefit in treating patients with metastatic hormone-sensitive PC with bisphosphonates without other medical indications for use. Additional studies suggest similar results for treatment with denosumab to that of bisphosphonate therapies.35
Nonmetastatic CRPC
Reasonable interest among treating clinicians exists to be able to delay or prevent the development of metastatic bone disease in patients who are showing biochemical signs of castration resistance but have not yet developed distant metastatic disease. Time to progression on ADT to castration resistance usually occurs 2 to 3 years following initiation of treatment. This typically occurs in patients with rising prostate-specific antigen (PSA). As per the Prostate Cancer Working Group 3, in the absence of radiologic progression, CRPC is defined by a 25% increase from the nadir (considering a starting value of ≥ 1 ng/mL), with a minimum rise of 2 ng/mL in the setting of castrate serum testosterone < 50 ng/dL despite good adherence to an ADT regimen, with proven serologic castration either by undetectable or a near undetectable nadir of serum testosterone concentration. Therapeutic implications include prevention of SREs as well as time to metastatic bone lesions. The Zometa 704 trial examined the use of ZA to reduce time to first metastatic bone lesion in the setting of patients with nonmetastatic CRPC.36 The trial was discontinued prematurely due to low patient accrual, but initial analysis provided information on the natural history of a rising PSA in this patient population. At 2 years, one-third of patients had developed bone metastases. Median bone metastasis-free survival was 30 months. Median time to first bone metastasis and OS were not reached. Baseline PSA and PSA velocity independently predicted a shorter time to first bone metastasis, metastasis-free survival, and OS.36
Denosumab was also studied in the setting of nonmetastatic CRPC in the Denosumab 147 trial. The study enrolled 1432 patients and found a significantly increased bone metastasis-free survival by a median of 4.2 months over placebo (HR, 0.85; 95% CI, 0.73-0.98; P = .03). Denosumab significantly delayed time to first bone metastasis (HR, 0.84; 95% CI, 0.71-0.98; P = .03). OS was similar between groups (HR, 1.01; 95% CI, 0.85-1.20; P = .91). Rates of AEs and serious AEs were similar between groups, except for ONJ and hypocalcemia. The rates of ONJ for denosumab were 1%, 3%, 4% in years 1,2, 3, respectively; overall, < 5% (n = 33). Hypocalcemia occurred in < 2% (n = 12) in denosumab-treated patients. The authors concluded that in men with CRPC, denosumab significantly prolonged bone metastasis–free survival and delayed time-to-bone metastasis.37 These 2 studies suggest a role of receptor-activated nuclear factor κ-B ligand inhibitor denosumab in patients with nonmetastatic CRPC in the appropriate setting. There were delays in bony metastatic disease, but no difference in OS. Rare denosumab treatment–related specific AEs were noted. Hence, denosumab is not recommended for use in this setting.
Metastatic CRPC
Castration resistance typically occurs 2 to 3 years following initiation of ADT and the most common extranodal site of disease is within the bone in metastatic PC. Disease progression within bones after ADT can be challenging given both the nature of progressive cancer with osteoblastic metastatic lesions and the prolonged effects of ADT on unaffected bone. The Zometa 039 study compared ZA with placebo and found a significant difference in SREs (38% and 49%, respectively; P .03). No survival benefit was observed with the addition of ZA. Use of other bisphosphonates pamidronate and clodronate did not have a similar degree of benefit.38,39
A phase 3 study of 1904 patients found that denosumab was superior to ZA in delaying the time to first on-study SRE (HR, 0.82; 95% CI, 0.71-0.95) and reducing rates of multiple SREs (HR, 0.82; 95% CI, 0.71-0.94).40 This was later confirmed with an additional study that demonstrated treatment with denosumab significantly reduced the risk of developing a first symptomatic SRE, defined as a pathologic fracture, spinal cord compression, necessity for radiation, or surgery (HR, 0.78; 95% CI, 0.66-0.93; P = .005) and first and subsequent symptomatic SREs (rate ratio, 0.78; 95% CI, 0.65-0.92; P = .004) compared with ZA.28 These findings suggest a continued role of denosumab in the treatment of advanced metastatic CRPC from both control of bone disease as well as quality of life and palliation of cancer-related symptoms.
Radium-223 dichloride (radium-223) is an α-emitting radionuclide for treatment of metastatic CRPC with bone metastasis, but otherwise no additional metastatic sites. Radium-223 is a calcium-mimetic that preferentially accumulates into areas of high-bone turnover, such as where bone metastases tend to occur. Radium-223 induces apoptosis of tumor cells through double-stranded DNA breaks. Studies have shown radium-223 to prolong OS and time-to-first symptomatic SRE.41 The ERA-223 trial showed that when radium-223 was combined with abiraterone acetate, there was an increase in fragility fracture risk compared with placebo combined with abiraterone. Data from the study revealed that the median symptomatic SRE-free survival was 22.3 months (95% CI, 20.4-24.8) in the radium-223 group and 26.0 months (21.8-28.3) in the placebo group. Concurrent treatment with abiraterone acetate plus prednisone or prednisolone and radium-223 was associated with increased fracture risk. Osteoporotic fractures were the most common type of fracture in the radium-223 group and of all fracture types, differed the most between the study groups.42
Conclusions
Convincing evidence supports the ongoing use of bisphosphonates and denosumab in patients with osteoporosis, significant osteopenia with risk factors, and in patients with CRPC with bone metastasis. Bone metastases can cause considerable morbidity and mortality among men with advanced PC. Pain, fracture, and neurologic injury can occur with metastatic bone lesions as well as with ADT-related bone loss. Prevention of SREs in patients with PC is a reasonable goal in PC survivors while being mindful of managing the risks of these therapies.
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7-30. doi:10.3322/caac.21590
2. Skolarus TA, Hawley ST. Prostate cancer survivorship care in the Veterans Health Administration. Fed Pract. 2014;31(8):10-17.
3. Gartrell BA, Coleman R, Efstathiou E, et al. Metastatic prostate cancer and the bone: significance and therapeutic options. Eur Urol. 2015;68(5):850-858. doi:10.1016/j.eururo.2015.06.039
4. Bolla M, de Reijke TM, Van Tienhoven G, et al. Duration of androgen suppression in the treatment of prostate cancer. N Engl J Med. 2009;360(24):2516-2527. doi:10.1056/NEJMoa0810095
5. Welch HG, Albertsen PC. Reconsidering Prostate cancer mortality—The future of PSA screening. N Engl J Med. 2020;382(16):1557-1563. doi:10.1056/NEJMms1914228
6. Coleman R, Body JJ, Aapro M, Hadji P, Herrstedt J; ESMO Guidelines Working Group. Bone health in cancer patients: ESMO Clinical Practice Guidelines. Ann Oncol. 2014;25 (suppl 3):iii124-137. doi:10.1093/annonc/mdu103
7. Saylor PJ, Smith MR. Adverse effects of androgen deprivation therapy: defining the problem and promoting health among men with prostate cancer. J Natl Compr Canc Netw. 2010;8(2):211-223. doi:10.6004/jnccn.2010.0014
8. Shahinian VB, Kuo Y-F, Freeman JL, Goodwin JS. Risk of fracture after androgen deprivation for prostate cancer. N Engl J Med. 2005;352(2):154-164. doi:10.1056/NEJMoa041943
9. Sartor O, de Bono JS. Metastatic prostate cancer. N Engl J Med. 2018;378(7):645-657. doi:10.1056/NEJMra1701695
10. Saad F, Eastham JA, Smith MR. Biochemical markers of bone turnover and clinical outcomes in men with prostate cancer. Urol Oncol. 2012;30(4):369-378. doi:10.1016/j.urolonc.2010.08.007
11. Cosman F, de Beur SJ, LeBoff MS, et al; National Osteoporosis Foundation. Clinician’s guide to prevention and treatment of osteoporosis. Osteoporos Int. 2014;25(10):2359-2381. doi:10.1007/s00198-014-2794-2
12. Alibhai SMH, Zukotynski K, Walker-Dilks C, et al; Cancer Care Ontario Genitourinary Cancer Disease Site Group. Bone health and bone-targeted therapies for prostate cancer: a programme in evidence-based care - Cancer Care Ontario Clinical Practice Guideline. Clin Oncol (R Coll Radiol). 2017;29(6):348-355. doi:10.1016/j.clon.2017.01.007
13. LEE CE. A comprehensive bone-health management approach with men with prostate cancer recieving androgen deprivation therapy. Curr Oncol. 2011;18(4):e163-172. doi:10.3747/co.v18i4.746
14. Kennel KA, Drake MT. Adverse effects of bisphosphonates: Implications for osteoporosis management. Mayo Clin Proc. 2009;84(7):632-638. doi:10.1016/S0025-6196(11)60752-0
15. Saad F, Brown JE, Van Poznak C, et al. Incidence, risk factors, and outcomes of osteonecrosis of the jaw: integrated analysis from three blinded active-controlled phase III trials in cancer patients with bone metastases. Ann Oncol. 2012;23(5):1341-1347. doi:10.1093/annonc/mdr435
16. Body J-J, Bone HG, de Boer RH, et al. Hypocalcaemia in patients with metastatic bone disease treated with denosumab. Eur J Cancer. 2015;51(13):1812-1821. doi:10.1016/j.ejca.2015.05.016
17. Wysowski DK, Chang JT. Alendronate and risedronate: reports of severe bone, joint, and muscle pain. Arch Intern Med. 2005;165(3):346-347. doi:10.1001/archinte.165.3.346-b
18. Saylor PJ, Rumble RB, Tagawa S, et al. Bone health and bone-targeted therapies for prostate cancer: ASCO endorsement of a cancer care Ontario guideline. J Clin Oncol. 2020;38(15):1736-1743. doi:10.1200/JCO.19.03148
19. Saad F, Gleason DM, Murray R, et al; Zoledronic Acid Prostate Cancer Study Group. Long-term efficacy of zoledronic acid for the prevention of skeletal complications in patients with metastatic hormone-refractory prostate cancer. J Natl Cancer Inst. 2004;96(11):879-882. doi:10.1093/jnci/djh141
20. Saad F, Gleason DM, Murray R, et al; Zoledronic Acid Prostate Cancer Study Group. A randomized, placebo-controlled trial of zoledronic zcid in patients with hormone-refractory metastatic prostate carcinoma. J Natl Cancer Inst. 2002;94(19):1458-1468. doi:10.1093/jnci/94.19.1458
21. Aapro M, Saad F. Bone-modifying agents in the treatment of bone metastases in patients with advanced genitourinary malignancies: a focus on zoledronic acid. Ther Adv Urol. 2012;4(2):85-101. doi:10.1177/1756287212441234
22. Cianferotti L, Bertoldo F, Carini M, et al. The prevention of fragility fractures in patients with non-metastatic prostate cancer: a position statement by the international osteoporosis foundation. Oncotarget. 2017;8(43):75646-75663. doi:10.18632/oncotarget.17980
23. Ruggiero S, Gralow J, Marx RE, et al. Practical guidelines for the prevention, diagnosis, and treatment of osteonecrosis of the jaw in patients with cancer. J Oncol Pract. 2006;2(1):7-14. doi:10.1200/JOP.2006.2.1.7
24. Corraini P, Heide-Jørgensen U, Schøodt M, et al. Osteonecrosis of the jaw and survival of patients with cancer: a nationwide cohort study in Denmark. Cancer Med. 2017;6(10):2271-2277. doi:10.1002/cam4.1173
25. Watts NB, Diab DL. Long-term use of bisphosphonates in osteoporosis. J Clin Endocrinol Metab. 2010;95(4):1555-1565. doi:10.1210/jc.2009-1947
26. Himelstein AL, Foster JC, Khatcheressian JL, et al. Effect of longer interval vs standard dosing of zoledronic acid on skeletal events in patients with bone metastases: a randomized clinical trial. JAMA. 2017;317(1):48-58. doi:10.1001/jama.2016.19425
27. Macherey S, Monsef I, Jahn F, et al. Bisphosphonates for advanced prostate cancer. Cochrane Database Syst Rev. 2017;12(12):CD006250. doi:10.1002/14651858.CD006250.pub2
28. Smith MR, Coleman RE, Klotz L, et al. Denosumab for the prevention of skeletal complications in metastatic castration-resistant prostate cancer: comparison of skeletal-related events and symptomatic skeletal events. Ann Oncol. 2015;26(2):368-374. doi:10.1093/annonc/mdu519
29. Wirth M, Tammela T, Cicalese V, et al. Prevention of bone metastases in patients with high-risk nonmetastatic prostate cancer treated with zoledronic acid: efficacy and safety results of the Zometa European Study (ZEUS). Eur Urol. 2015;67(3):482-491. doi:10.1016/j.eururo.2014.02.014
30. James ND, Sydes MR, Clarke NW, et al; STAMPEDE Investigators. Addition of docetaxel, zoledronic acid, or both to first-line long-term hormone therapy in prostate cancer (STAMPEDE): survival results from an adaptive, multiarm, multistage, platform randomised controlled trial. Lancet. 2016;387(10024):1163-1177. doi:10.1016/S0140-6736(15)01037-5
31. Denham JW, Joseph D, Lamb DS, et al. Short-term androgen suppression and radiotherapy versus intermediate-term androgen suppression and radiotherapy, with or without zoledronic acid, in men with locally advanced prostate cancer (TROG 03.04 RADAR): 10-year results from a randomised, phase 3, factorial trial. Lancet Oncol. 2019;20(2):267-281. doi:10.1016/S1470-2045(18)30757-5
32. Dearnaley DP, Mason MD, Parmar MK, Sanders K, Sydes MR. Adjuvant therapy with oral sodium clodronate in locally advanced and metastatic prostate cancer: long-term overall survival results from the MRC PR04 and PR05 randomised controlled trials. Lancet Oncol. 2009;10(9):872-876. doi:10.1016/S1470-2045(09)70201-3
33. Smith MR, Egerdie B, Toriz NH, et al; Denosumab HALT Prostate Cancer Study Group. Denosumab in men receiving androgen-deprivation therapy for prostate Cancer. N Engl J Med. 2009;361(8):745-755. doi:10.1056/NEJMoa0809003
34. Smith MR, Halabi S, Ryan CJ, et al. Randomized controlled trial of early zoledronic acid in men with castration-sensitive prostate cancer and bone metastases: results of CALGB 90202 (alliance). J Clin Oncol. 2014;32(11):1143-1150. doi:10.1200/JCO.2013.51.6500
35. Kozyrakis D, Paridis D, Perikleous S, Malizos K, Zarkadas A, Tsagkalis A. The current role of osteoclast inhibitors in patients with prostate cancer. Adv Urol. 2018;2018:1525832. doi:10.1155/2018/1525832
36. Smith MR, Kabbinavar F, Saad F, et al. Natural history of rising serum prostate-specific antigen in men with castrate nonmetastatic prostate cancer. J Clin Oncol. 2005;23(13):2918-2925. doi:10.1200/JCO.2005.01.529
37. Smith MR, Saad F, Coleman R, et al. Denosumab and bone-metastasis-free survival in men with castration-resistant prostate cancer: results of a phase 3, randomised, placebo-controlled trial. Lancet. 2012;379(9810):39-46. doi:10.1016/S0140-6736(11)61226-9
38. Small EJ, Smith MR, Seaman JJ, Petrone S, Kowalski MO. Combined analysis of two multicenter, randomized, placebo-controlled studies of pamidronate disodium for the palliation of bone pain in men with metastatic prostate cancer. J Clin Oncol. 2003;21(23):4277-4284. doi:10.1200/JCO.2003.05.147
39. Ernst DS, Tannock IF, Winquist EW, et al. Randomized, double-blind, controlled trial of mitoxantrone/prednisone and clodronate versus mitoxantrone/prednisone and placebo in patients with hormone-refractory prostate cancer and pain. J Clin Oncol. 2003;21(17):3335-3342. doi:10.1200/JCO.2003.03.042
40. Fizazi K, Carducci M, Smith M, et al. Denosumab versus zoledronic acid for treatment of bone metastases in men with castration-resistant prostate cancer: a randomised, double-blind study. Lancet. 2011;377(9768):813-822. doi:10.1016/S0140-6736(10)62344-6
41. Parker C, Nilsson S, Heinrich D, et al; ALSYMPCA Investigators Alpha emitter radium-223 and survival in metastatic prostate cancer. N Engl J Med. 2013;369(3):213-223. doi:10.1056/NEJMoa1213755
42. Smith M, Parker C, Saad F, et al. Addition of radium-223 to abiraterone acetate and prednisone or prednisolone in patients with castration-resistant prostate cancer and bone metastases (ERA 223): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2019;20(3):408-419. doi:10.1016/S1470-2045(18)30860-X
43. Smith MR, Saad F, Shore ND, et al. Effect of denosumab on prolonging bone-metastasis-free survival (BMFS) in men with nonmetastatic castrate-resistant prostate cancer (CRPC) presenting with aggressive PSA kinetics. J Clin Oncol. 2012;30(5_suppl):6-6.
44. Saad F, Gleason DM, Murray R, et al; Zoledronic Acid Prostate Cancer Study Group. A randomized, placebo-controlled trial of zoledronic acid in patients with hormone-refractory metastatic prostate carcinoma. J Natl Cancer Inst. 2002;94(19):1458-1468. doi:10.1093/jnci/94.19.1458
Prostate cancer (PC) is the most commonly and newly diagnosed nonskin cancer and the second leading cause of cancer death in men in the United States. About 191,930 cases and about 33,330 deaths from PC were expected for the year 2020.1 About 1 in 41 men will die of PC. Most men diagnosed with PC are aged > 65 years and do not die of their disease. The 5-year survival rate of localized and regional disease is nearly 100%, and disease with distant metastases is 31%. As a result, more than 3.1 million men in the United States who have been diagnosed with PC are still alive today.1 Among veterans, there is a substantial population living with PC. Skolarus and Hawley reported in 2014 that an estimated 200,000 veterans with PC were survivors and 12,000 were newly diagnosed.2
In PC, skeletal strength can be affected by several factors, such as aging, malnutrition, androgen-deprivation therapy (ADT), and bone metastasis.3,4 In fact, most men can live the rest of their life with PC by using strategies to monitor and treat it, once it shows either radiographic or chemical signs of progression.5 ADT is the standard of care to treat hormone-sensitive PC, which is associated with significant skeletal-related adverse effects (AEs).6,7
Men undergoing ADT are 4 times more likely to develop substantial bone deficiency, Shahinian and colleagues found that in men surviving 5 years after PC diagnosis, 19.4% of those who received ADT had a fracture compared with 12% in men who did not (P < .001). The authors established a significant relation between the number of doses of gonadotropin-releasing hormone given in the first 12 months and the risk of fracture.8 Of those who progressed to metastatic disease, the first metastatic nonnodal site is most commonly to the bone.9 Advanced PC is characterized by increased bone turnover, which further raises concerns for bone health and patient performance.10
Skeletal-related events (SREs) include pathologic fracture, spinal cord compression, palliative radiation, or surgery to bone, and change in antineoplastic therapy secondary to bone pain. The concept of bone health refers to the prevention, diagnosis, and treatment of idiopathic, pathogenic, and treatment-related bone loss and delay or prevention of SREs.6,11 Guidelines and expert groups have recommended screening for osteoporosis at the start of ADT with bone mineral density testing, ensuring adequate calcium and vitamin D intake, modifying lifestyle behaviors (smoking cessation, alcohol moderation, and regular exercise), and prescribing bisphosphonates or receptor-activated nuclear factor κ-B ligand inhibitor, denosumab, for men with osteoporosis or who are at general high-fracture risk.12,13 The overuse of these medications results in undue cost to patients as well as AEs, such as osteonecrosis of the jaw (ONJ), hypocalcemia, and bone/joint pains.14-17 There are evidence-based guidelines for appropriate use of bisphosphonates and denosumab for delay and prevention of SREs in the setting of advanced PC.18 These doses also typically differ in frequency to those of osteoporosis.19 We summarize the evidence and guidance for health care providers who care for patients with PC at various stages and complications from both disease-related and treatment-related comorbidities.
Bone-Strengthening Agents
Overall, there is evidence to support the use of bone-strengthening agents in patients with osteopenia/osteoporosis in the prevention of SREs with significant risk factors for progressive bone demineralization, such as lifestyle factors and, in particular, treatments such as ADT. Bone-remodeling agents for treatment of bony metastasis have been shown to provide therapeutic advantage only in limited instances in the castration-resistant PC (CRPC) setting. Hence, in patients with hormone-sensitive PC due to medication-related AEs, treatment with bone-strengthening agents is indicated only if the patient has a significant preexisting risk for fracture from osteopenia/osteoporosis (Table). The Figure depicts an algorithm for the management of bone health in men with PC who are being treated with ADT.
Denosumab and bisphosphonates have an established role in preventing SREs in metastatic CRPC.20 The choice of denosumab or a bisphosphonate typically varies based on the indication, possible AEs, and cost of therapy. There are multiple studies involving initiation of these agents at various stages of disease to improve both time to progression as well as management of SREs. There is a lack of evidence that bisphosphonates prevent metastatic-bone lesions in castration-sensitive PC; therefore, prophylactic use of this agent is not recommended in patients unless they have significant bone demineralization.21,22
Medication-induced ONJ is a severe AE of both denosumab and bisphosphonate therapies. Data from recent trials showed that higher dosing and prolonged duration of denosumab and bisphosphonate therapies further increased risk of ONJ by 1.8% and 1.3%, respectively.15 Careful history taking and discussions with the patient and if possible their dentist on how to reduce risk are recommended. It is good practice for the patient to complete a dental evaluation prior to starting IV bisphosphonates or denosumab. Dental evaluations should be performed routinely at 3- to 12-month intervals throughout therapy based on individualized risk assessment.23 The benefits of using bisphosphonates to prevent fractures associated with osteoporosis outweigh the risk of ONJ in high-risk populations, but not in all patients with PC. A case-by-case basis and evaluation of risk factors should be performed prior to administering bone-modifying therapy. The long-term safety of IV bisphosphonates has not been adequately studied in controlled trials, and concerns regarding long-term complications, including renal toxicity, ONJ, and atypical femoral fractures, remain with prolonged therapy.24,25
The CALGB 70604 (Alliance) trial compared 3-month dosing to monthly treatment with zoledronic acid (ZA), showing no inferiority to lower frequency dosing.26 A Cochrane review of clinical trials found that in patients with advanced PC, bisphosphonates were found to provide roughly 58 fewer SREs per 1000 on average.27 A phase 3 study showed a modest benefit to denosumab vs ZA in the CRPC group regarding incidence of SREs. The rates of SREs were 289 of 951 patients in the bisphosphonate group, and 241 of 950 patients in the denosumab group (30.4% vs 25.3%; hazard ratio [HR], 0.78; 95% CI, 0.66-0.93; P = .005).28 In 2020, the American Society of Clinical Oncology endorsed the Cancer Care Ontario guidelines for prostate bone health care.18 Adequate supplementation is necessary in all patients treated with a bisphosphonate or denosumab to prevent treatment-related hypocalcemia. Typically, daily supplementation with a minimum of calcium 500 mg and vitamin D 400 IU is recommended.16
Bone Health in Patients
Nonmetastatic Hormone-Sensitive PC
ADT forms the backbone of treatment for patients with local and advanced metastatic castration-sensitive PC along with surgical and focal radiotherapy options. Cancer treatment-induced bone loss is known to occur with prolonged use of ADT. The ZEUS trial found no prevention of bone metastasis in patients with high-risk localized PC with the use of ZA in the absence of bone metastasis. A Kaplan-Meier estimated proportion of bone metastases after a median follow-up of 4.8 years was found to be not statistically significant: 14.7% in the ZA group vs 13.2% in the control/placebo group.29 The STAMPEDE trial showed no significant overall survival (OS) benefit with the addition of ZA to ADT vs ADT alone (HR, 0.94; 95% CI, 0.79-1.11; P = .45), 5-year survival with ADT alone was 55% compared to ADT plus ZA with 57% 5-year survival.30 The RADAR trial showed that at 5 years in high Gleason score patients, use of ZA in the absence of bone metastasis was beneficial, but not in low- or intermediate-risk patients. However, at 10-year analysis there was no significant difference in any of the high-stratified groups with or without ZA.31
The PR04 trial showed no effect on OS with clodronate compared with placebo in nonmetastatic castration-sensitive PC, with a HR of 1.12 (95% CI, 0.89-1.42; P = .94). The estimated 5-year survival was 80% with placebo and 78% with clodronate; 10-year survival rates were 51% with placebo and 48% with clodronate.32 Data from the HALT trial showed an increased bone mineral density and reduced risk of new vertebral fractures vs placebo (1.5% vs 3.9%, respectively) in the absence of metastatic bone lesions and a reduction in new vertebral fractures in patients with nonmetastatic PC.33 Most of these studies showed no benefit with the addition of ZA to nonmetastatic PC; although, the HALT trial provides evidence to support use of denosumab in patients with nonmetastatic PC for preventing vertebral fragility fractures in men receiving ADT.
Metastatic Hormone-Sensitive PC
ZA is often used to treat men with metastatic castration-sensitive PC despite limited efficacy and safety data. The CALGB 90202 (Alliance) trial authors found that the early use of ZA was not associated with increased time to first SRE. The median time to first SRE was 31.9 months in the ZA group (95% CI, 24.2-40.3) and 29.8 months in the placebo group (stratified HR, 0.97; 95% CI, 0-1.17; 1-sided stratified log-rank P = .39).34 OS was similar between the groups (HR, 0.88; 95% CI, 0.70-1.12; P = .29) as were reported AEs.34 Results from these studies suggest limited benefit in treating patients with metastatic hormone-sensitive PC with bisphosphonates without other medical indications for use. Additional studies suggest similar results for treatment with denosumab to that of bisphosphonate therapies.35
Nonmetastatic CRPC
Reasonable interest among treating clinicians exists to be able to delay or prevent the development of metastatic bone disease in patients who are showing biochemical signs of castration resistance but have not yet developed distant metastatic disease. Time to progression on ADT to castration resistance usually occurs 2 to 3 years following initiation of treatment. This typically occurs in patients with rising prostate-specific antigen (PSA). As per the Prostate Cancer Working Group 3, in the absence of radiologic progression, CRPC is defined by a 25% increase from the nadir (considering a starting value of ≥ 1 ng/mL), with a minimum rise of 2 ng/mL in the setting of castrate serum testosterone < 50 ng/dL despite good adherence to an ADT regimen, with proven serologic castration either by undetectable or a near undetectable nadir of serum testosterone concentration. Therapeutic implications include prevention of SREs as well as time to metastatic bone lesions. The Zometa 704 trial examined the use of ZA to reduce time to first metastatic bone lesion in the setting of patients with nonmetastatic CRPC.36 The trial was discontinued prematurely due to low patient accrual, but initial analysis provided information on the natural history of a rising PSA in this patient population. At 2 years, one-third of patients had developed bone metastases. Median bone metastasis-free survival was 30 months. Median time to first bone metastasis and OS were not reached. Baseline PSA and PSA velocity independently predicted a shorter time to first bone metastasis, metastasis-free survival, and OS.36
Denosumab was also studied in the setting of nonmetastatic CRPC in the Denosumab 147 trial. The study enrolled 1432 patients and found a significantly increased bone metastasis-free survival by a median of 4.2 months over placebo (HR, 0.85; 95% CI, 0.73-0.98; P = .03). Denosumab significantly delayed time to first bone metastasis (HR, 0.84; 95% CI, 0.71-0.98; P = .03). OS was similar between groups (HR, 1.01; 95% CI, 0.85-1.20; P = .91). Rates of AEs and serious AEs were similar between groups, except for ONJ and hypocalcemia. The rates of ONJ for denosumab were 1%, 3%, 4% in years 1,2, 3, respectively; overall, < 5% (n = 33). Hypocalcemia occurred in < 2% (n = 12) in denosumab-treated patients. The authors concluded that in men with CRPC, denosumab significantly prolonged bone metastasis–free survival and delayed time-to-bone metastasis.37 These 2 studies suggest a role of receptor-activated nuclear factor κ-B ligand inhibitor denosumab in patients with nonmetastatic CRPC in the appropriate setting. There were delays in bony metastatic disease, but no difference in OS. Rare denosumab treatment–related specific AEs were noted. Hence, denosumab is not recommended for use in this setting.
Metastatic CRPC
Castration resistance typically occurs 2 to 3 years following initiation of ADT and the most common extranodal site of disease is within the bone in metastatic PC. Disease progression within bones after ADT can be challenging given both the nature of progressive cancer with osteoblastic metastatic lesions and the prolonged effects of ADT on unaffected bone. The Zometa 039 study compared ZA with placebo and found a significant difference in SREs (38% and 49%, respectively; P .03). No survival benefit was observed with the addition of ZA. Use of other bisphosphonates pamidronate and clodronate did not have a similar degree of benefit.38,39
A phase 3 study of 1904 patients found that denosumab was superior to ZA in delaying the time to first on-study SRE (HR, 0.82; 95% CI, 0.71-0.95) and reducing rates of multiple SREs (HR, 0.82; 95% CI, 0.71-0.94).40 This was later confirmed with an additional study that demonstrated treatment with denosumab significantly reduced the risk of developing a first symptomatic SRE, defined as a pathologic fracture, spinal cord compression, necessity for radiation, or surgery (HR, 0.78; 95% CI, 0.66-0.93; P = .005) and first and subsequent symptomatic SREs (rate ratio, 0.78; 95% CI, 0.65-0.92; P = .004) compared with ZA.28 These findings suggest a continued role of denosumab in the treatment of advanced metastatic CRPC from both control of bone disease as well as quality of life and palliation of cancer-related symptoms.
Radium-223 dichloride (radium-223) is an α-emitting radionuclide for treatment of metastatic CRPC with bone metastasis, but otherwise no additional metastatic sites. Radium-223 is a calcium-mimetic that preferentially accumulates into areas of high-bone turnover, such as where bone metastases tend to occur. Radium-223 induces apoptosis of tumor cells through double-stranded DNA breaks. Studies have shown radium-223 to prolong OS and time-to-first symptomatic SRE.41 The ERA-223 trial showed that when radium-223 was combined with abiraterone acetate, there was an increase in fragility fracture risk compared with placebo combined with abiraterone. Data from the study revealed that the median symptomatic SRE-free survival was 22.3 months (95% CI, 20.4-24.8) in the radium-223 group and 26.0 months (21.8-28.3) in the placebo group. Concurrent treatment with abiraterone acetate plus prednisone or prednisolone and radium-223 was associated with increased fracture risk. Osteoporotic fractures were the most common type of fracture in the radium-223 group and of all fracture types, differed the most between the study groups.42
Conclusions
Convincing evidence supports the ongoing use of bisphosphonates and denosumab in patients with osteoporosis, significant osteopenia with risk factors, and in patients with CRPC with bone metastasis. Bone metastases can cause considerable morbidity and mortality among men with advanced PC. Pain, fracture, and neurologic injury can occur with metastatic bone lesions as well as with ADT-related bone loss. Prevention of SREs in patients with PC is a reasonable goal in PC survivors while being mindful of managing the risks of these therapies.
Prostate cancer (PC) is the most commonly and newly diagnosed nonskin cancer and the second leading cause of cancer death in men in the United States. About 191,930 cases and about 33,330 deaths from PC were expected for the year 2020.1 About 1 in 41 men will die of PC. Most men diagnosed with PC are aged > 65 years and do not die of their disease. The 5-year survival rate of localized and regional disease is nearly 100%, and disease with distant metastases is 31%. As a result, more than 3.1 million men in the United States who have been diagnosed with PC are still alive today.1 Among veterans, there is a substantial population living with PC. Skolarus and Hawley reported in 2014 that an estimated 200,000 veterans with PC were survivors and 12,000 were newly diagnosed.2
In PC, skeletal strength can be affected by several factors, such as aging, malnutrition, androgen-deprivation therapy (ADT), and bone metastasis.3,4 In fact, most men can live the rest of their life with PC by using strategies to monitor and treat it, once it shows either radiographic or chemical signs of progression.5 ADT is the standard of care to treat hormone-sensitive PC, which is associated with significant skeletal-related adverse effects (AEs).6,7
Men undergoing ADT are 4 times more likely to develop substantial bone deficiency, Shahinian and colleagues found that in men surviving 5 years after PC diagnosis, 19.4% of those who received ADT had a fracture compared with 12% in men who did not (P < .001). The authors established a significant relation between the number of doses of gonadotropin-releasing hormone given in the first 12 months and the risk of fracture.8 Of those who progressed to metastatic disease, the first metastatic nonnodal site is most commonly to the bone.9 Advanced PC is characterized by increased bone turnover, which further raises concerns for bone health and patient performance.10
Skeletal-related events (SREs) include pathologic fracture, spinal cord compression, palliative radiation, or surgery to bone, and change in antineoplastic therapy secondary to bone pain. The concept of bone health refers to the prevention, diagnosis, and treatment of idiopathic, pathogenic, and treatment-related bone loss and delay or prevention of SREs.6,11 Guidelines and expert groups have recommended screening for osteoporosis at the start of ADT with bone mineral density testing, ensuring adequate calcium and vitamin D intake, modifying lifestyle behaviors (smoking cessation, alcohol moderation, and regular exercise), and prescribing bisphosphonates or receptor-activated nuclear factor κ-B ligand inhibitor, denosumab, for men with osteoporosis or who are at general high-fracture risk.12,13 The overuse of these medications results in undue cost to patients as well as AEs, such as osteonecrosis of the jaw (ONJ), hypocalcemia, and bone/joint pains.14-17 There are evidence-based guidelines for appropriate use of bisphosphonates and denosumab for delay and prevention of SREs in the setting of advanced PC.18 These doses also typically differ in frequency to those of osteoporosis.19 We summarize the evidence and guidance for health care providers who care for patients with PC at various stages and complications from both disease-related and treatment-related comorbidities.
Bone-Strengthening Agents
Overall, there is evidence to support the use of bone-strengthening agents in patients with osteopenia/osteoporosis in the prevention of SREs with significant risk factors for progressive bone demineralization, such as lifestyle factors and, in particular, treatments such as ADT. Bone-remodeling agents for treatment of bony metastasis have been shown to provide therapeutic advantage only in limited instances in the castration-resistant PC (CRPC) setting. Hence, in patients with hormone-sensitive PC due to medication-related AEs, treatment with bone-strengthening agents is indicated only if the patient has a significant preexisting risk for fracture from osteopenia/osteoporosis (Table). The Figure depicts an algorithm for the management of bone health in men with PC who are being treated with ADT.
Denosumab and bisphosphonates have an established role in preventing SREs in metastatic CRPC.20 The choice of denosumab or a bisphosphonate typically varies based on the indication, possible AEs, and cost of therapy. There are multiple studies involving initiation of these agents at various stages of disease to improve both time to progression as well as management of SREs. There is a lack of evidence that bisphosphonates prevent metastatic-bone lesions in castration-sensitive PC; therefore, prophylactic use of this agent is not recommended in patients unless they have significant bone demineralization.21,22
Medication-induced ONJ is a severe AE of both denosumab and bisphosphonate therapies. Data from recent trials showed that higher dosing and prolonged duration of denosumab and bisphosphonate therapies further increased risk of ONJ by 1.8% and 1.3%, respectively.15 Careful history taking and discussions with the patient and if possible their dentist on how to reduce risk are recommended. It is good practice for the patient to complete a dental evaluation prior to starting IV bisphosphonates or denosumab. Dental evaluations should be performed routinely at 3- to 12-month intervals throughout therapy based on individualized risk assessment.23 The benefits of using bisphosphonates to prevent fractures associated with osteoporosis outweigh the risk of ONJ in high-risk populations, but not in all patients with PC. A case-by-case basis and evaluation of risk factors should be performed prior to administering bone-modifying therapy. The long-term safety of IV bisphosphonates has not been adequately studied in controlled trials, and concerns regarding long-term complications, including renal toxicity, ONJ, and atypical femoral fractures, remain with prolonged therapy.24,25
The CALGB 70604 (Alliance) trial compared 3-month dosing to monthly treatment with zoledronic acid (ZA), showing no inferiority to lower frequency dosing.26 A Cochrane review of clinical trials found that in patients with advanced PC, bisphosphonates were found to provide roughly 58 fewer SREs per 1000 on average.27 A phase 3 study showed a modest benefit to denosumab vs ZA in the CRPC group regarding incidence of SREs. The rates of SREs were 289 of 951 patients in the bisphosphonate group, and 241 of 950 patients in the denosumab group (30.4% vs 25.3%; hazard ratio [HR], 0.78; 95% CI, 0.66-0.93; P = .005).28 In 2020, the American Society of Clinical Oncology endorsed the Cancer Care Ontario guidelines for prostate bone health care.18 Adequate supplementation is necessary in all patients treated with a bisphosphonate or denosumab to prevent treatment-related hypocalcemia. Typically, daily supplementation with a minimum of calcium 500 mg and vitamin D 400 IU is recommended.16
Bone Health in Patients
Nonmetastatic Hormone-Sensitive PC
ADT forms the backbone of treatment for patients with local and advanced metastatic castration-sensitive PC along with surgical and focal radiotherapy options. Cancer treatment-induced bone loss is known to occur with prolonged use of ADT. The ZEUS trial found no prevention of bone metastasis in patients with high-risk localized PC with the use of ZA in the absence of bone metastasis. A Kaplan-Meier estimated proportion of bone metastases after a median follow-up of 4.8 years was found to be not statistically significant: 14.7% in the ZA group vs 13.2% in the control/placebo group.29 The STAMPEDE trial showed no significant overall survival (OS) benefit with the addition of ZA to ADT vs ADT alone (HR, 0.94; 95% CI, 0.79-1.11; P = .45), 5-year survival with ADT alone was 55% compared to ADT plus ZA with 57% 5-year survival.30 The RADAR trial showed that at 5 years in high Gleason score patients, use of ZA in the absence of bone metastasis was beneficial, but not in low- or intermediate-risk patients. However, at 10-year analysis there was no significant difference in any of the high-stratified groups with or without ZA.31
The PR04 trial showed no effect on OS with clodronate compared with placebo in nonmetastatic castration-sensitive PC, with a HR of 1.12 (95% CI, 0.89-1.42; P = .94). The estimated 5-year survival was 80% with placebo and 78% with clodronate; 10-year survival rates were 51% with placebo and 48% with clodronate.32 Data from the HALT trial showed an increased bone mineral density and reduced risk of new vertebral fractures vs placebo (1.5% vs 3.9%, respectively) in the absence of metastatic bone lesions and a reduction in new vertebral fractures in patients with nonmetastatic PC.33 Most of these studies showed no benefit with the addition of ZA to nonmetastatic PC; although, the HALT trial provides evidence to support use of denosumab in patients with nonmetastatic PC for preventing vertebral fragility fractures in men receiving ADT.
Metastatic Hormone-Sensitive PC
ZA is often used to treat men with metastatic castration-sensitive PC despite limited efficacy and safety data. The CALGB 90202 (Alliance) trial authors found that the early use of ZA was not associated with increased time to first SRE. The median time to first SRE was 31.9 months in the ZA group (95% CI, 24.2-40.3) and 29.8 months in the placebo group (stratified HR, 0.97; 95% CI, 0-1.17; 1-sided stratified log-rank P = .39).34 OS was similar between the groups (HR, 0.88; 95% CI, 0.70-1.12; P = .29) as were reported AEs.34 Results from these studies suggest limited benefit in treating patients with metastatic hormone-sensitive PC with bisphosphonates without other medical indications for use. Additional studies suggest similar results for treatment with denosumab to that of bisphosphonate therapies.35
Nonmetastatic CRPC
Reasonable interest among treating clinicians exists to be able to delay or prevent the development of metastatic bone disease in patients who are showing biochemical signs of castration resistance but have not yet developed distant metastatic disease. Time to progression on ADT to castration resistance usually occurs 2 to 3 years following initiation of treatment. This typically occurs in patients with rising prostate-specific antigen (PSA). As per the Prostate Cancer Working Group 3, in the absence of radiologic progression, CRPC is defined by a 25% increase from the nadir (considering a starting value of ≥ 1 ng/mL), with a minimum rise of 2 ng/mL in the setting of castrate serum testosterone < 50 ng/dL despite good adherence to an ADT regimen, with proven serologic castration either by undetectable or a near undetectable nadir of serum testosterone concentration. Therapeutic implications include prevention of SREs as well as time to metastatic bone lesions. The Zometa 704 trial examined the use of ZA to reduce time to first metastatic bone lesion in the setting of patients with nonmetastatic CRPC.36 The trial was discontinued prematurely due to low patient accrual, but initial analysis provided information on the natural history of a rising PSA in this patient population. At 2 years, one-third of patients had developed bone metastases. Median bone metastasis-free survival was 30 months. Median time to first bone metastasis and OS were not reached. Baseline PSA and PSA velocity independently predicted a shorter time to first bone metastasis, metastasis-free survival, and OS.36
Denosumab was also studied in the setting of nonmetastatic CRPC in the Denosumab 147 trial. The study enrolled 1432 patients and found a significantly increased bone metastasis-free survival by a median of 4.2 months over placebo (HR, 0.85; 95% CI, 0.73-0.98; P = .03). Denosumab significantly delayed time to first bone metastasis (HR, 0.84; 95% CI, 0.71-0.98; P = .03). OS was similar between groups (HR, 1.01; 95% CI, 0.85-1.20; P = .91). Rates of AEs and serious AEs were similar between groups, except for ONJ and hypocalcemia. The rates of ONJ for denosumab were 1%, 3%, 4% in years 1,2, 3, respectively; overall, < 5% (n = 33). Hypocalcemia occurred in < 2% (n = 12) in denosumab-treated patients. The authors concluded that in men with CRPC, denosumab significantly prolonged bone metastasis–free survival and delayed time-to-bone metastasis.37 These 2 studies suggest a role of receptor-activated nuclear factor κ-B ligand inhibitor denosumab in patients with nonmetastatic CRPC in the appropriate setting. There were delays in bony metastatic disease, but no difference in OS. Rare denosumab treatment–related specific AEs were noted. Hence, denosumab is not recommended for use in this setting.
Metastatic CRPC
Castration resistance typically occurs 2 to 3 years following initiation of ADT and the most common extranodal site of disease is within the bone in metastatic PC. Disease progression within bones after ADT can be challenging given both the nature of progressive cancer with osteoblastic metastatic lesions and the prolonged effects of ADT on unaffected bone. The Zometa 039 study compared ZA with placebo and found a significant difference in SREs (38% and 49%, respectively; P .03). No survival benefit was observed with the addition of ZA. Use of other bisphosphonates pamidronate and clodronate did not have a similar degree of benefit.38,39
A phase 3 study of 1904 patients found that denosumab was superior to ZA in delaying the time to first on-study SRE (HR, 0.82; 95% CI, 0.71-0.95) and reducing rates of multiple SREs (HR, 0.82; 95% CI, 0.71-0.94).40 This was later confirmed with an additional study that demonstrated treatment with denosumab significantly reduced the risk of developing a first symptomatic SRE, defined as a pathologic fracture, spinal cord compression, necessity for radiation, or surgery (HR, 0.78; 95% CI, 0.66-0.93; P = .005) and first and subsequent symptomatic SREs (rate ratio, 0.78; 95% CI, 0.65-0.92; P = .004) compared with ZA.28 These findings suggest a continued role of denosumab in the treatment of advanced metastatic CRPC from both control of bone disease as well as quality of life and palliation of cancer-related symptoms.
Radium-223 dichloride (radium-223) is an α-emitting radionuclide for treatment of metastatic CRPC with bone metastasis, but otherwise no additional metastatic sites. Radium-223 is a calcium-mimetic that preferentially accumulates into areas of high-bone turnover, such as where bone metastases tend to occur. Radium-223 induces apoptosis of tumor cells through double-stranded DNA breaks. Studies have shown radium-223 to prolong OS and time-to-first symptomatic SRE.41 The ERA-223 trial showed that when radium-223 was combined with abiraterone acetate, there was an increase in fragility fracture risk compared with placebo combined with abiraterone. Data from the study revealed that the median symptomatic SRE-free survival was 22.3 months (95% CI, 20.4-24.8) in the radium-223 group and 26.0 months (21.8-28.3) in the placebo group. Concurrent treatment with abiraterone acetate plus prednisone or prednisolone and radium-223 was associated with increased fracture risk. Osteoporotic fractures were the most common type of fracture in the radium-223 group and of all fracture types, differed the most between the study groups.42
Conclusions
Convincing evidence supports the ongoing use of bisphosphonates and denosumab in patients with osteoporosis, significant osteopenia with risk factors, and in patients with CRPC with bone metastasis. Bone metastases can cause considerable morbidity and mortality among men with advanced PC. Pain, fracture, and neurologic injury can occur with metastatic bone lesions as well as with ADT-related bone loss. Prevention of SREs in patients with PC is a reasonable goal in PC survivors while being mindful of managing the risks of these therapies.
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7-30. doi:10.3322/caac.21590
2. Skolarus TA, Hawley ST. Prostate cancer survivorship care in the Veterans Health Administration. Fed Pract. 2014;31(8):10-17.
3. Gartrell BA, Coleman R, Efstathiou E, et al. Metastatic prostate cancer and the bone: significance and therapeutic options. Eur Urol. 2015;68(5):850-858. doi:10.1016/j.eururo.2015.06.039
4. Bolla M, de Reijke TM, Van Tienhoven G, et al. Duration of androgen suppression in the treatment of prostate cancer. N Engl J Med. 2009;360(24):2516-2527. doi:10.1056/NEJMoa0810095
5. Welch HG, Albertsen PC. Reconsidering Prostate cancer mortality—The future of PSA screening. N Engl J Med. 2020;382(16):1557-1563. doi:10.1056/NEJMms1914228
6. Coleman R, Body JJ, Aapro M, Hadji P, Herrstedt J; ESMO Guidelines Working Group. Bone health in cancer patients: ESMO Clinical Practice Guidelines. Ann Oncol. 2014;25 (suppl 3):iii124-137. doi:10.1093/annonc/mdu103
7. Saylor PJ, Smith MR. Adverse effects of androgen deprivation therapy: defining the problem and promoting health among men with prostate cancer. J Natl Compr Canc Netw. 2010;8(2):211-223. doi:10.6004/jnccn.2010.0014
8. Shahinian VB, Kuo Y-F, Freeman JL, Goodwin JS. Risk of fracture after androgen deprivation for prostate cancer. N Engl J Med. 2005;352(2):154-164. doi:10.1056/NEJMoa041943
9. Sartor O, de Bono JS. Metastatic prostate cancer. N Engl J Med. 2018;378(7):645-657. doi:10.1056/NEJMra1701695
10. Saad F, Eastham JA, Smith MR. Biochemical markers of bone turnover and clinical outcomes in men with prostate cancer. Urol Oncol. 2012;30(4):369-378. doi:10.1016/j.urolonc.2010.08.007
11. Cosman F, de Beur SJ, LeBoff MS, et al; National Osteoporosis Foundation. Clinician’s guide to prevention and treatment of osteoporosis. Osteoporos Int. 2014;25(10):2359-2381. doi:10.1007/s00198-014-2794-2
12. Alibhai SMH, Zukotynski K, Walker-Dilks C, et al; Cancer Care Ontario Genitourinary Cancer Disease Site Group. Bone health and bone-targeted therapies for prostate cancer: a programme in evidence-based care - Cancer Care Ontario Clinical Practice Guideline. Clin Oncol (R Coll Radiol). 2017;29(6):348-355. doi:10.1016/j.clon.2017.01.007
13. LEE CE. A comprehensive bone-health management approach with men with prostate cancer recieving androgen deprivation therapy. Curr Oncol. 2011;18(4):e163-172. doi:10.3747/co.v18i4.746
14. Kennel KA, Drake MT. Adverse effects of bisphosphonates: Implications for osteoporosis management. Mayo Clin Proc. 2009;84(7):632-638. doi:10.1016/S0025-6196(11)60752-0
15. Saad F, Brown JE, Van Poznak C, et al. Incidence, risk factors, and outcomes of osteonecrosis of the jaw: integrated analysis from three blinded active-controlled phase III trials in cancer patients with bone metastases. Ann Oncol. 2012;23(5):1341-1347. doi:10.1093/annonc/mdr435
16. Body J-J, Bone HG, de Boer RH, et al. Hypocalcaemia in patients with metastatic bone disease treated with denosumab. Eur J Cancer. 2015;51(13):1812-1821. doi:10.1016/j.ejca.2015.05.016
17. Wysowski DK, Chang JT. Alendronate and risedronate: reports of severe bone, joint, and muscle pain. Arch Intern Med. 2005;165(3):346-347. doi:10.1001/archinte.165.3.346-b
18. Saylor PJ, Rumble RB, Tagawa S, et al. Bone health and bone-targeted therapies for prostate cancer: ASCO endorsement of a cancer care Ontario guideline. J Clin Oncol. 2020;38(15):1736-1743. doi:10.1200/JCO.19.03148
19. Saad F, Gleason DM, Murray R, et al; Zoledronic Acid Prostate Cancer Study Group. Long-term efficacy of zoledronic acid for the prevention of skeletal complications in patients with metastatic hormone-refractory prostate cancer. J Natl Cancer Inst. 2004;96(11):879-882. doi:10.1093/jnci/djh141
20. Saad F, Gleason DM, Murray R, et al; Zoledronic Acid Prostate Cancer Study Group. A randomized, placebo-controlled trial of zoledronic zcid in patients with hormone-refractory metastatic prostate carcinoma. J Natl Cancer Inst. 2002;94(19):1458-1468. doi:10.1093/jnci/94.19.1458
21. Aapro M, Saad F. Bone-modifying agents in the treatment of bone metastases in patients with advanced genitourinary malignancies: a focus on zoledronic acid. Ther Adv Urol. 2012;4(2):85-101. doi:10.1177/1756287212441234
22. Cianferotti L, Bertoldo F, Carini M, et al. The prevention of fragility fractures in patients with non-metastatic prostate cancer: a position statement by the international osteoporosis foundation. Oncotarget. 2017;8(43):75646-75663. doi:10.18632/oncotarget.17980
23. Ruggiero S, Gralow J, Marx RE, et al. Practical guidelines for the prevention, diagnosis, and treatment of osteonecrosis of the jaw in patients with cancer. J Oncol Pract. 2006;2(1):7-14. doi:10.1200/JOP.2006.2.1.7
24. Corraini P, Heide-Jørgensen U, Schøodt M, et al. Osteonecrosis of the jaw and survival of patients with cancer: a nationwide cohort study in Denmark. Cancer Med. 2017;6(10):2271-2277. doi:10.1002/cam4.1173
25. Watts NB, Diab DL. Long-term use of bisphosphonates in osteoporosis. J Clin Endocrinol Metab. 2010;95(4):1555-1565. doi:10.1210/jc.2009-1947
26. Himelstein AL, Foster JC, Khatcheressian JL, et al. Effect of longer interval vs standard dosing of zoledronic acid on skeletal events in patients with bone metastases: a randomized clinical trial. JAMA. 2017;317(1):48-58. doi:10.1001/jama.2016.19425
27. Macherey S, Monsef I, Jahn F, et al. Bisphosphonates for advanced prostate cancer. Cochrane Database Syst Rev. 2017;12(12):CD006250. doi:10.1002/14651858.CD006250.pub2
28. Smith MR, Coleman RE, Klotz L, et al. Denosumab for the prevention of skeletal complications in metastatic castration-resistant prostate cancer: comparison of skeletal-related events and symptomatic skeletal events. Ann Oncol. 2015;26(2):368-374. doi:10.1093/annonc/mdu519
29. Wirth M, Tammela T, Cicalese V, et al. Prevention of bone metastases in patients with high-risk nonmetastatic prostate cancer treated with zoledronic acid: efficacy and safety results of the Zometa European Study (ZEUS). Eur Urol. 2015;67(3):482-491. doi:10.1016/j.eururo.2014.02.014
30. James ND, Sydes MR, Clarke NW, et al; STAMPEDE Investigators. Addition of docetaxel, zoledronic acid, or both to first-line long-term hormone therapy in prostate cancer (STAMPEDE): survival results from an adaptive, multiarm, multistage, platform randomised controlled trial. Lancet. 2016;387(10024):1163-1177. doi:10.1016/S0140-6736(15)01037-5
31. Denham JW, Joseph D, Lamb DS, et al. Short-term androgen suppression and radiotherapy versus intermediate-term androgen suppression and radiotherapy, with or without zoledronic acid, in men with locally advanced prostate cancer (TROG 03.04 RADAR): 10-year results from a randomised, phase 3, factorial trial. Lancet Oncol. 2019;20(2):267-281. doi:10.1016/S1470-2045(18)30757-5
32. Dearnaley DP, Mason MD, Parmar MK, Sanders K, Sydes MR. Adjuvant therapy with oral sodium clodronate in locally advanced and metastatic prostate cancer: long-term overall survival results from the MRC PR04 and PR05 randomised controlled trials. Lancet Oncol. 2009;10(9):872-876. doi:10.1016/S1470-2045(09)70201-3
33. Smith MR, Egerdie B, Toriz NH, et al; Denosumab HALT Prostate Cancer Study Group. Denosumab in men receiving androgen-deprivation therapy for prostate Cancer. N Engl J Med. 2009;361(8):745-755. doi:10.1056/NEJMoa0809003
34. Smith MR, Halabi S, Ryan CJ, et al. Randomized controlled trial of early zoledronic acid in men with castration-sensitive prostate cancer and bone metastases: results of CALGB 90202 (alliance). J Clin Oncol. 2014;32(11):1143-1150. doi:10.1200/JCO.2013.51.6500
35. Kozyrakis D, Paridis D, Perikleous S, Malizos K, Zarkadas A, Tsagkalis A. The current role of osteoclast inhibitors in patients with prostate cancer. Adv Urol. 2018;2018:1525832. doi:10.1155/2018/1525832
36. Smith MR, Kabbinavar F, Saad F, et al. Natural history of rising serum prostate-specific antigen in men with castrate nonmetastatic prostate cancer. J Clin Oncol. 2005;23(13):2918-2925. doi:10.1200/JCO.2005.01.529
37. Smith MR, Saad F, Coleman R, et al. Denosumab and bone-metastasis-free survival in men with castration-resistant prostate cancer: results of a phase 3, randomised, placebo-controlled trial. Lancet. 2012;379(9810):39-46. doi:10.1016/S0140-6736(11)61226-9
38. Small EJ, Smith MR, Seaman JJ, Petrone S, Kowalski MO. Combined analysis of two multicenter, randomized, placebo-controlled studies of pamidronate disodium for the palliation of bone pain in men with metastatic prostate cancer. J Clin Oncol. 2003;21(23):4277-4284. doi:10.1200/JCO.2003.05.147
39. Ernst DS, Tannock IF, Winquist EW, et al. Randomized, double-blind, controlled trial of mitoxantrone/prednisone and clodronate versus mitoxantrone/prednisone and placebo in patients with hormone-refractory prostate cancer and pain. J Clin Oncol. 2003;21(17):3335-3342. doi:10.1200/JCO.2003.03.042
40. Fizazi K, Carducci M, Smith M, et al. Denosumab versus zoledronic acid for treatment of bone metastases in men with castration-resistant prostate cancer: a randomised, double-blind study. Lancet. 2011;377(9768):813-822. doi:10.1016/S0140-6736(10)62344-6
41. Parker C, Nilsson S, Heinrich D, et al; ALSYMPCA Investigators Alpha emitter radium-223 and survival in metastatic prostate cancer. N Engl J Med. 2013;369(3):213-223. doi:10.1056/NEJMoa1213755
42. Smith M, Parker C, Saad F, et al. Addition of radium-223 to abiraterone acetate and prednisone or prednisolone in patients with castration-resistant prostate cancer and bone metastases (ERA 223): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2019;20(3):408-419. doi:10.1016/S1470-2045(18)30860-X
43. Smith MR, Saad F, Shore ND, et al. Effect of denosumab on prolonging bone-metastasis-free survival (BMFS) in men with nonmetastatic castrate-resistant prostate cancer (CRPC) presenting with aggressive PSA kinetics. J Clin Oncol. 2012;30(5_suppl):6-6.
44. Saad F, Gleason DM, Murray R, et al; Zoledronic Acid Prostate Cancer Study Group. A randomized, placebo-controlled trial of zoledronic acid in patients with hormone-refractory metastatic prostate carcinoma. J Natl Cancer Inst. 2002;94(19):1458-1468. doi:10.1093/jnci/94.19.1458
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7-30. doi:10.3322/caac.21590
2. Skolarus TA, Hawley ST. Prostate cancer survivorship care in the Veterans Health Administration. Fed Pract. 2014;31(8):10-17.
3. Gartrell BA, Coleman R, Efstathiou E, et al. Metastatic prostate cancer and the bone: significance and therapeutic options. Eur Urol. 2015;68(5):850-858. doi:10.1016/j.eururo.2015.06.039
4. Bolla M, de Reijke TM, Van Tienhoven G, et al. Duration of androgen suppression in the treatment of prostate cancer. N Engl J Med. 2009;360(24):2516-2527. doi:10.1056/NEJMoa0810095
5. Welch HG, Albertsen PC. Reconsidering Prostate cancer mortality—The future of PSA screening. N Engl J Med. 2020;382(16):1557-1563. doi:10.1056/NEJMms1914228
6. Coleman R, Body JJ, Aapro M, Hadji P, Herrstedt J; ESMO Guidelines Working Group. Bone health in cancer patients: ESMO Clinical Practice Guidelines. Ann Oncol. 2014;25 (suppl 3):iii124-137. doi:10.1093/annonc/mdu103
7. Saylor PJ, Smith MR. Adverse effects of androgen deprivation therapy: defining the problem and promoting health among men with prostate cancer. J Natl Compr Canc Netw. 2010;8(2):211-223. doi:10.6004/jnccn.2010.0014
8. Shahinian VB, Kuo Y-F, Freeman JL, Goodwin JS. Risk of fracture after androgen deprivation for prostate cancer. N Engl J Med. 2005;352(2):154-164. doi:10.1056/NEJMoa041943
9. Sartor O, de Bono JS. Metastatic prostate cancer. N Engl J Med. 2018;378(7):645-657. doi:10.1056/NEJMra1701695
10. Saad F, Eastham JA, Smith MR. Biochemical markers of bone turnover and clinical outcomes in men with prostate cancer. Urol Oncol. 2012;30(4):369-378. doi:10.1016/j.urolonc.2010.08.007
11. Cosman F, de Beur SJ, LeBoff MS, et al; National Osteoporosis Foundation. Clinician’s guide to prevention and treatment of osteoporosis. Osteoporos Int. 2014;25(10):2359-2381. doi:10.1007/s00198-014-2794-2
12. Alibhai SMH, Zukotynski K, Walker-Dilks C, et al; Cancer Care Ontario Genitourinary Cancer Disease Site Group. Bone health and bone-targeted therapies for prostate cancer: a programme in evidence-based care - Cancer Care Ontario Clinical Practice Guideline. Clin Oncol (R Coll Radiol). 2017;29(6):348-355. doi:10.1016/j.clon.2017.01.007
13. LEE CE. A comprehensive bone-health management approach with men with prostate cancer recieving androgen deprivation therapy. Curr Oncol. 2011;18(4):e163-172. doi:10.3747/co.v18i4.746
14. Kennel KA, Drake MT. Adverse effects of bisphosphonates: Implications for osteoporosis management. Mayo Clin Proc. 2009;84(7):632-638. doi:10.1016/S0025-6196(11)60752-0
15. Saad F, Brown JE, Van Poznak C, et al. Incidence, risk factors, and outcomes of osteonecrosis of the jaw: integrated analysis from three blinded active-controlled phase III trials in cancer patients with bone metastases. Ann Oncol. 2012;23(5):1341-1347. doi:10.1093/annonc/mdr435
16. Body J-J, Bone HG, de Boer RH, et al. Hypocalcaemia in patients with metastatic bone disease treated with denosumab. Eur J Cancer. 2015;51(13):1812-1821. doi:10.1016/j.ejca.2015.05.016
17. Wysowski DK, Chang JT. Alendronate and risedronate: reports of severe bone, joint, and muscle pain. Arch Intern Med. 2005;165(3):346-347. doi:10.1001/archinte.165.3.346-b
18. Saylor PJ, Rumble RB, Tagawa S, et al. Bone health and bone-targeted therapies for prostate cancer: ASCO endorsement of a cancer care Ontario guideline. J Clin Oncol. 2020;38(15):1736-1743. doi:10.1200/JCO.19.03148
19. Saad F, Gleason DM, Murray R, et al; Zoledronic Acid Prostate Cancer Study Group. Long-term efficacy of zoledronic acid for the prevention of skeletal complications in patients with metastatic hormone-refractory prostate cancer. J Natl Cancer Inst. 2004;96(11):879-882. doi:10.1093/jnci/djh141
20. Saad F, Gleason DM, Murray R, et al; Zoledronic Acid Prostate Cancer Study Group. A randomized, placebo-controlled trial of zoledronic zcid in patients with hormone-refractory metastatic prostate carcinoma. J Natl Cancer Inst. 2002;94(19):1458-1468. doi:10.1093/jnci/94.19.1458
21. Aapro M, Saad F. Bone-modifying agents in the treatment of bone metastases in patients with advanced genitourinary malignancies: a focus on zoledronic acid. Ther Adv Urol. 2012;4(2):85-101. doi:10.1177/1756287212441234
22. Cianferotti L, Bertoldo F, Carini M, et al. The prevention of fragility fractures in patients with non-metastatic prostate cancer: a position statement by the international osteoporosis foundation. Oncotarget. 2017;8(43):75646-75663. doi:10.18632/oncotarget.17980
23. Ruggiero S, Gralow J, Marx RE, et al. Practical guidelines for the prevention, diagnosis, and treatment of osteonecrosis of the jaw in patients with cancer. J Oncol Pract. 2006;2(1):7-14. doi:10.1200/JOP.2006.2.1.7
24. Corraini P, Heide-Jørgensen U, Schøodt M, et al. Osteonecrosis of the jaw and survival of patients with cancer: a nationwide cohort study in Denmark. Cancer Med. 2017;6(10):2271-2277. doi:10.1002/cam4.1173
25. Watts NB, Diab DL. Long-term use of bisphosphonates in osteoporosis. J Clin Endocrinol Metab. 2010;95(4):1555-1565. doi:10.1210/jc.2009-1947
26. Himelstein AL, Foster JC, Khatcheressian JL, et al. Effect of longer interval vs standard dosing of zoledronic acid on skeletal events in patients with bone metastases: a randomized clinical trial. JAMA. 2017;317(1):48-58. doi:10.1001/jama.2016.19425
27. Macherey S, Monsef I, Jahn F, et al. Bisphosphonates for advanced prostate cancer. Cochrane Database Syst Rev. 2017;12(12):CD006250. doi:10.1002/14651858.CD006250.pub2
28. Smith MR, Coleman RE, Klotz L, et al. Denosumab for the prevention of skeletal complications in metastatic castration-resistant prostate cancer: comparison of skeletal-related events and symptomatic skeletal events. Ann Oncol. 2015;26(2):368-374. doi:10.1093/annonc/mdu519
29. Wirth M, Tammela T, Cicalese V, et al. Prevention of bone metastases in patients with high-risk nonmetastatic prostate cancer treated with zoledronic acid: efficacy and safety results of the Zometa European Study (ZEUS). Eur Urol. 2015;67(3):482-491. doi:10.1016/j.eururo.2014.02.014
30. James ND, Sydes MR, Clarke NW, et al; STAMPEDE Investigators. Addition of docetaxel, zoledronic acid, or both to first-line long-term hormone therapy in prostate cancer (STAMPEDE): survival results from an adaptive, multiarm, multistage, platform randomised controlled trial. Lancet. 2016;387(10024):1163-1177. doi:10.1016/S0140-6736(15)01037-5
31. Denham JW, Joseph D, Lamb DS, et al. Short-term androgen suppression and radiotherapy versus intermediate-term androgen suppression and radiotherapy, with or without zoledronic acid, in men with locally advanced prostate cancer (TROG 03.04 RADAR): 10-year results from a randomised, phase 3, factorial trial. Lancet Oncol. 2019;20(2):267-281. doi:10.1016/S1470-2045(18)30757-5
32. Dearnaley DP, Mason MD, Parmar MK, Sanders K, Sydes MR. Adjuvant therapy with oral sodium clodronate in locally advanced and metastatic prostate cancer: long-term overall survival results from the MRC PR04 and PR05 randomised controlled trials. Lancet Oncol. 2009;10(9):872-876. doi:10.1016/S1470-2045(09)70201-3
33. Smith MR, Egerdie B, Toriz NH, et al; Denosumab HALT Prostate Cancer Study Group. Denosumab in men receiving androgen-deprivation therapy for prostate Cancer. N Engl J Med. 2009;361(8):745-755. doi:10.1056/NEJMoa0809003
34. Smith MR, Halabi S, Ryan CJ, et al. Randomized controlled trial of early zoledronic acid in men with castration-sensitive prostate cancer and bone metastases: results of CALGB 90202 (alliance). J Clin Oncol. 2014;32(11):1143-1150. doi:10.1200/JCO.2013.51.6500
35. Kozyrakis D, Paridis D, Perikleous S, Malizos K, Zarkadas A, Tsagkalis A. The current role of osteoclast inhibitors in patients with prostate cancer. Adv Urol. 2018;2018:1525832. doi:10.1155/2018/1525832
36. Smith MR, Kabbinavar F, Saad F, et al. Natural history of rising serum prostate-specific antigen in men with castrate nonmetastatic prostate cancer. J Clin Oncol. 2005;23(13):2918-2925. doi:10.1200/JCO.2005.01.529
37. Smith MR, Saad F, Coleman R, et al. Denosumab and bone-metastasis-free survival in men with castration-resistant prostate cancer: results of a phase 3, randomised, placebo-controlled trial. Lancet. 2012;379(9810):39-46. doi:10.1016/S0140-6736(11)61226-9
38. Small EJ, Smith MR, Seaman JJ, Petrone S, Kowalski MO. Combined analysis of two multicenter, randomized, placebo-controlled studies of pamidronate disodium for the palliation of bone pain in men with metastatic prostate cancer. J Clin Oncol. 2003;21(23):4277-4284. doi:10.1200/JCO.2003.05.147
39. Ernst DS, Tannock IF, Winquist EW, et al. Randomized, double-blind, controlled trial of mitoxantrone/prednisone and clodronate versus mitoxantrone/prednisone and placebo in patients with hormone-refractory prostate cancer and pain. J Clin Oncol. 2003;21(17):3335-3342. doi:10.1200/JCO.2003.03.042
40. Fizazi K, Carducci M, Smith M, et al. Denosumab versus zoledronic acid for treatment of bone metastases in men with castration-resistant prostate cancer: a randomised, double-blind study. Lancet. 2011;377(9768):813-822. doi:10.1016/S0140-6736(10)62344-6
41. Parker C, Nilsson S, Heinrich D, et al; ALSYMPCA Investigators Alpha emitter radium-223 and survival in metastatic prostate cancer. N Engl J Med. 2013;369(3):213-223. doi:10.1056/NEJMoa1213755
42. Smith M, Parker C, Saad F, et al. Addition of radium-223 to abiraterone acetate and prednisone or prednisolone in patients with castration-resistant prostate cancer and bone metastases (ERA 223): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2019;20(3):408-419. doi:10.1016/S1470-2045(18)30860-X
43. Smith MR, Saad F, Shore ND, et al. Effect of denosumab on prolonging bone-metastasis-free survival (BMFS) in men with nonmetastatic castrate-resistant prostate cancer (CRPC) presenting with aggressive PSA kinetics. J Clin Oncol. 2012;30(5_suppl):6-6.
44. Saad F, Gleason DM, Murray R, et al; Zoledronic Acid Prostate Cancer Study Group. A randomized, placebo-controlled trial of zoledronic acid in patients with hormone-refractory metastatic prostate carcinoma. J Natl Cancer Inst. 2002;94(19):1458-1468. doi:10.1093/jnci/94.19.1458
An Interdisciplinary Approach to Metastatic Pancreatic Cancer and Comorbid Opioid Use Disorder Treatment Within a VA Health Care System
A multidisciplinary approach provided safe and feasible cancer treatment in a patient with advanced pancreatic cancer and coexisting active substance use disorder.
Substance use disorders (SUDs) are an important but understudied aspect of treating patients diagnosed with cancer. Substance use can affect cancer treatment outcomes, including morbidity and mortality.1,2 Additionally, patients with cancer and SUD may have unique psychosocial needs that require close attention and management. There is a paucity of data regarding the best approach to treating such patients. For example, cocaine use may increase the cardiovascular and hematologic risk of some traditional chemotherapy agents.3,4 Newer targeted agents and immunotherapies remain understudied with respect to SUD risk.
Although the US Department of Veterans Affairs (VA) has established helpful clinical practice guidelines for the treatment of SUD, there are no guidelines for treating patients with SUD and cancer.5 Clinicians have limited confidence in treatment approach, and treatment is inconsistent among oncologists nationwide even within the same practice. Furthermore, it can be challenging to safely prescribe opioids for cancer-related pain in individuals with SUD. There is a high risk of SUD and mental health disorders in veterans, making this population particularly vulnerable. We report a case of a male with metastatic pancreatic cancer, severe opioid use disorder (OUD) and moderate cocaine use disorder (CUD) who received pain management and cancer treatment under the direction of a multidisciplinary team approach.
Case Report
A 63-year-old male with a medical history of HIV treated with highly active antiretroviral therapy (HAART), compensated cirrhosis, severe OUD, moderate CUD, and sedative use disorder in sustained remission was admitted to the West Haven campus of the VA Connecticut Healthcare System (VACHS) with abdominal pain, weight loss and fatigue. He used heroin 1 month prior to his admission and reported regular cocaine and marijuana use (Table 1). He was diagnosed with HIV in 1989, and his medical history included herpes zoster and oral candidiasis but no other opportunistic infections. Several months prior to this admission, he had an undetectable viral load and CD4 count of 688.
At the time of this admission, the patient was adherent to methadone treatment. He reported increased abdominal pain. Computed tomography (CT) showed a 2.4-cm mass in the pancreatic uncinate process, multiple liver metastases, retroperitoneal lymphadenopathy, and small lung nodules. A CT-guided liver biopsy showed adenocarcinoma consistent with a primary cancer of the pancreas. Given the complexity of the case, a multidisciplinary team approach was used to treat his cancer and the sequelae safely, including the oncology team, community living center team, palliative care team, and interprofessional opioid reassessment clinic team (ORC).
Cancer Treatment
Chemotherapy with FOLFIRINOX (leucovorin calcium, fluorouracil, irinotecan hydrochloride, and oxaliplatin) was recommended. The first cycle of treatment originally was planned for the outpatient setting, and a peripherally inserted central catheter (PICC) line was placed. However, after a urine toxicology test was positive for cocaine, the PICC line was removed due to concern for possible use of PICC line for nonprescribed substance use. The patient expressed suicidal ideation at the time and was admitted for psychiatric consult and pain control. Cycle 1 FOLFIRINOX was started during this admission. A PICC line was again put in place and then removed before discharge. A celiac plexus block was performed several days after this admission for pain control.
Given concern about cocaine use increasing the risk of cardiac toxicity with FOLFIRINOX treatment, treating providers sconsulted with the community living center (CLC) about possible admission for future chemotherapy administration and pain management. The CLC at VACHS has 38 beds for rehabilitation, long-term care, and hospice with the mission to restore each veteran to his or her highest level of well-being. After discussion with this patient and CLC staff, he agreed to a CLC admission. The patient agreed to remain in the facility, wear a secure care device, and not leave without staff accompaniment. He was able to obtain a 2-hour pass to pay bills and rent. During the 2 months he was admitted to the CLC he would present to the VACHS Cancer Center for chemotherapy every 2 weeks. He completed 6 cycles of chemotherapy while admitted. During the admission, he was transferred to active medical service for 2 days for fever and malaise, and then returned to the CLC. The patient elected to leave the CLC after 2 months as the inability to see close friends was interfering with his quality of life.
Upon being discharged from the CLC, shared decision making took place with the patient to establish a new treatment plan. In collaboration with the patient, a plan was made to admit him every 2 weeks for continued chemotherapy. A PICC line was placed on each day of admission and removed prior to discharge. It was also agreed that treatment would be delayed if a urine drug test was positive for cocaine on the morning of admission. The patient was also seen by ORC every 2 weeks after being discharged from the CLC.
Imaging after cycle 6 showed decreased size of liver metastases, retroperitoneal lymph nodes, and pancreas mass. Cancer antigen 19-9 (CA19-9) tumor marker was reduced from 3513 U/mL pretreatment to 50 U/mL after cycle 7. Chemotherapy cycle 7 was delayed 6 days due to active cocaine and heroin use. A repeat urine was obtained several days later, which was negative for cocaine, and he was admitted for cycle 7 chemotherapy. Using this treatment approach of admissions for every cycle, the patient was able to receive 11 cycles of FOLFIRINOX with clinical benefit.
Palliative Care/Pain Management
Safely treating the patient’s malignant pain in the context of his OUD was critically important. In order to do this the palliative care team worked closely alongside ORC, is a multidisciplinary team consisting of health care providers (HCPs) from addiction psychiatry, internal medicine, health psychology and pharmacy who are consulted to evaluate veterans’ current opioid regimens and make recommendations to optimize both safety and efficacy. ORC followed this particular veteran as an outpatient and consulted on pain issues during his admission. They recommended the continuation of methadone at 120 mg daily and increased oral oxycodone to 30 mg every 6 hours, and then further increased to 45 mg every 6 hours. He continued to have increased pain despite higher doses of oxycodone, and pain medication was changed to oral hydromorphone 28 mg every 6 hours with the continuation of methadone. ORC and the palliative care team obtained consent from the veteran and a release of Information form signed by the patient to contact his community methadone clinic for further collaboration around pain management throughout the time caring for the veteran.
Even with improvement in disease based on imaging and tumor markers, opioid medications could not be decreased in this case. This is likely in part due to the multidimensional nature of pain. Careful assessment of the biologic, emotional, social, and spiritual contributors to pain is needed in the management of pain, especially at end of life.6 Nonpharmacologic pain management strategies used in this case included a transcutaneous electrical nerve stimulation unit, moist heat, celiac plexus block, and emotional support.
Psychosocial Issues/Substance Use
Psychosocial support for the patient was provided by the interdisciplinary palliative care team and the ORC team in both the inpatient and outpatient settings. Despite efforts from case management to get the veteran home services once discharged from the CLC, he declined repeatedly. Thus, the CLC social worker obtained a guardian alert for the veteran on discharge.
Close outpatient follow-up for medical and psychosocial support was very critical. When an outpatient, the veteran was scheduled for biweekly appointments with palliative care or ORC. When admitted to the hospital, the palliative care team medical director and psychologist conducted joint visits with him. Although he denied depressed mood and anxiety throughout his treatment, he often reflected on regrets that he had as he faced the end of his life. Specifically, he shared thoughts about being estranged from his surviving brother given his long struggle with substance use. Although he did not think a relationship was possible with his brother at the end of life, he still cared deeply for him and wanted to make him aware of his pancreatic cancer diagnosis. This was particularly important to him because their late brother had also died of pancreatic cancer. It was the patient’s wish at the end of his life to alert his surviving brother of his diagnosis so he and his children could get adequate screening throughout their lives. Although he had spoken of this desire often, it wasn’t until his disease progressed and he elected to transition to hospice that he felt ready to write the letter. The palliative care team assisted the veteran in writing and mailing a letter to his brother informing him of his diagnosis and transition to hospice as well as communicating that his brother and his family had been in his thoughts at the end of his life. The patient’s brother received this letter and with assistance from the CLC social worker made arrangements to visit the veteran at bedside at the inpatient CLC hospice unit the final days of his life.
Discussion
There are very little data on the safety of cancer-directed therapy in patients with active SUD. The limited studies that have been done showed conflicting results.
A retrospective study among women with co-occurring SUD and locally advanced cervical cancer who were undergoing primary radiation therapy found that SUD was not associated with a difference in toxicity or survival outcomes.7 However, other research suggests that SUD may be associated with an increase in all-cause mortality as well as other adverse outcomes for patients and health care systems (eg, emergency department visits, hospitalizations).8 A retrospective study of patients with a history of SUD and nonsmall cell lung cancer showed that these patients had higher rates of depression, less family support, increased rates of missed appointments, more emergency department visits and more hospitalizations.9 Patients with chronic myeloid leukemia or myelodysplastic syndromes who had long-term cocaine use had a 6-fold increased risk of death, which was not found in patients who had long-term alcohol or marijuana use.2
The limited data highlight the need for careful consideration of ways to mitigate potentially adverse outcomes in this population while still providing clinically indicated cancer treatment. Integrated VA health care systems provide unique resources that can maximize veteran safety during cancer treatment. Utilization of VA resources and close interdisciplinary collaboration across VA HCPs can help to ensure equitable access to state-of-the-art cancer therapies for veterans with comorbid SUD.
VA Services for Patients With Comorbidities
This case highlights several distinct aspects of VA health care that make it possible to safely treat individuals with complex comorbidities. One important aspect of this was collaboration with the CLC to admit the veteran for his initial treatment after a positive cocaine test. CLC admission was nonpunitive and allowed ongoing involvement in the VA community. This provided an essential, safe, and structured environment in which 6 cycles of chemotherapy could be delivered.
Although the patient left the CLC after 2 months due to floor restrictions negatively impacting his quality of life and ability to spend time with close friends, several important events occurred during this stay. First, the patient established close relationships with the CLC staff and the palliative care team; both groups followed him throughout his inpatient and outpatient care. These relationships proved essential throughout his care as they were the foundation of difficult conversations about substance use, treatment adherence, and eventually, transition to hospice.
In addition, the opportunity to administer 6 cycles of chemotherapy at the CLC was enough to lead to clinical benefit and radiographic response to treatment. Clinical benefits while in the CLC included maintenance of a good appetite, 15-lb weight gain and preserved performance status (ECOG [Eastern Cooperative Group]-1), which allowed him to actively participate in multiple social and recreational activities while in the CLC. From early conversations, this patient was clear that he wanted treatment as long as his life could be prolonged with good quality of life. Having evidence of the benefit of treatment, at least initially, increased the patient’s confidence in treatment. There were a few conversations when the challenges of treatment mounted (eg, pain, needs for abstinence from cocaine prior to admission for chemotherapy, frequent doctor appointments), and the patient would remind himself of these data to recommit himself to treatment. The opportunity to admit him to the inpatient VA facility, including bed availability for 3 days during his treatment once he left the CLC was important. This plan to admit the patient following a negative urine toxicology test for cocaine was made collaboratively with the veteran and the oncology and palliative care teams. The plan allowed the patient to achieve his treatment goals while maintaining his safety and reducing theoretical cardiac toxicities with his cancer treatment.
Finally, the availability of a multidisciplinary team approach including palliative care, oncology, psychology, addiction medicine and addiction psychiatry, was critical for addressing the veteran’s malignant pain. Palliative care worked in close collaboration with the ORC to prescribe and renew pain medications. ORC offered ongoing consultation on pain management in the context of OUD. As the veteran’s cancer progressed and functional decline prohibited his daily attendance at the community methadone clinic, palliative care and ORC met with the methadone clinic to arrange a less frequent methadone pickup schedule (the patient previously needed daily pickup). Non-VA settings may not have access to these resources to safely treat the biopsychosocial issues that arise in complex cases.
Substance Use and Cancer Treatments
This case raises several critical questions for oncologic care. Cocaine and fluorouracil are both associated with cardiotoxicity, and many oncologists would not feel it is safe to administer a regimen containing fluorouracil to a patient with active cocaine use. The National Comprehensive Cancer Network (NCCN) panel recommends FOLFIRINOX as a preferred category 1 recommendation for first-line treatment of patients with advanced pancreas cancer with good performance status.10 This recommendation is based on the PRODIGE trial, which has shown improved overall survival (OS): 11.1 vs 6.8 months for patients who received single-agent gemcitabine.11 If patients are not candidates for FOLFIRINOX and have good performance status, the NCCN recommends gemcitabine plus albumin-bound paclitaxel with category 1 level of evidence based on the IMPACT trial, which showed improvement in OS (8.7 vs 6.6 months compared with single-agent gemcitabine).12
Some oncologists may have additional concerns administering fluorouracil treatment alternatives (such as gemcitabine and albumin-bound paclitaxel) to individuals with active SUD because of concerns about altered mental status impacting the ability to report important adverse effects. In the absence of sufficient data, HCPs must determine whether they feel it is safe to administer these agents in individuals with active cocaine use. However, denying these patients the possible benefits of standard-of-care life-prolonging therapies without established data raises concerns regarding the ethics of such practices. There is concern that the stigma surrounding cocaine use might contribute to withholding treatment, while treatment is continued for individuals taking prescribed stimulant medications that also have cardiotoxicity risks. VA health care facilities are uniquely situated to use all available resources to address these issues using interprofessional patient-centered care and determine the most optimal treatment based on a risk/benefit discussion between the patient and the HCP.
Similarly, this case also raised questions among HCPs about the safety of using an indwelling port for treatment in a patient with SUD. In the current case there was concern about keeping in a port for a patient with a history of IV drug use; therefore, a PICC line was initiated and removed at each admission. Without guidelines in these situations, HCPs are left to weigh the risks and benefits of using a port or a PICC for individuals with recent or current substance use without formal data, which can lead to inconsistent access to care. More guidance is needed for these situations.
SUD Screening
This case begs the question of whether oncologists are adequately screening for a range of SUDs, and when they encounter an issue, how they are addressing it. Many oncologists do not receive adequate training on assessment of current or recent substance use. There are health care and systems-level practices that may increase patient safety for individuals with ongoing substance use who are undergoing cancer treatment. Training on obtaining appropriate substance use histories, motivational interviewing to resolve ambivalence about substance use in the direction of change, and shared decision making about treatment options could increase confidence in understanding and addressing substance use issues. It is also important to educate oncologists on how to address patients who return to or continued substance use during treatment. In this case the collaboration from palliative care, psychology, addiction medicine, and addiction psychiatry through the ORC was essential in assisting with ongoing assessment of substance use, guiding difficult conversations about the impact of substance use on the treatment plan, and identifying risk-mitigation strategies. Close collaboration and full utilization of all VA resources allowed this patient to receive first-line treatment for pancreatic cancer in order to reach his goal of prolonging his life while maintaining acceptable quality of life. Table 2 provides best practices for management of patients with comorbid SUD and cancer.
More research is needed into cancer treatment for patients with SUD, especially in the current era of cancer care using novel cancer treatments leading to significantly improved survival in many cancer types. Ideally, oncologists should be routinely or consistently screening patients for substance use, including alcohol. The patient should participate in this decision-making process after being educated about the risks and benefits. These patients can be followed using a multimodal approach to increase their rates of success and improve their quality of life. Although the literature is limited and no formal guidelines are available, VA oncologists are fortunate to have a range of resources available to them to navigate these difficult cases. Veterans have elevated rates of SUD, making this a critical issue to consider in the VA.13 It is the hope that this case can highlight how to take advantage of the many VA resources in order to ensure equitable cancer care for all veterans.
Conclusions
This case demonstrates that cancer-directed treatment is safe and feasible in a patient with advanced pancreatic cancer and coexisting active SUD by using a multidisciplinary approach. The multidisciplinary team included palliative care, oncology, psychology, addiction medicine, and addiction psychiatry. Critical steps for a successful outcome include gathering history about SUD; motivational interviewing to resolve ambivalence about treatment for SUD; shared decision making about cancer treatment; and risk-reduction strategies in pain and SUD management.
Treatment advancements in many cancer types have led to significantly longer survival, and it is critical to develop safe protocols to treat patients with active SUD so they also can derive benefit from these very significant medical advancements.
Acknowledgments
Michal Rose, MD, Director of VACHS Cancer Center, and Chandrika Kumar, MD, Director of VACHS Community Living Center, for their collaboration in care for this veteran.
1. Chang G, Meadows ME, Jones JA, Antin JH, Orav EJ. Substance use and survival after treatment for chronic myelogenous leukemia (CML) or myelodysplastic syndrome (MDS). Am J Drug Alcohol Ab. 2010;36(1):1-6. doi:10.3109/00952990903490758
2. Stagno S, Busby K, Shapiro A, Kotz M. Patients at risk: addressing addiction in patients undergoing hematopoietic SCT. Bone Marrow Transplant. 2008;42(4):221-226. doi:10.1038/bmt.2008.211
3. Arora NP. Cutaneous vasculopathy and neutropenia associated with levamisole-adulterated cocaine. Am J Med Sci. 2013;345(1):45-51. doi:10.1097/MAJ.0b013e31825b2b50
4. Schwartz BG, Rezkalla S, Kloner RA. Cardiovascular effects of cocaine. Circulation. 2010;122(24):2558-2569. doi:10.1161/CIRCULATIONAHA.110.940569
5. US Department of Veterans Affairs, US Department of Defense. VA/DoD clinical practice guideline for the management of substance use disorders. Published 2015. Accessed July 8, 2021. https://www.healthquality.va.gov/guidelines/MH/sud/VADODSUDCPGRevised22216.pdf
6. Mehta A, Chan LS. Understanding of the concept of “total pain”: a prerequisite for pain control. J Hosp Palliat Nurs. 2008;10(1):26-32. doi:10.1097/01.NJH.0000306714.50539.1a
7. Rubinsak LA, Terplan M, Martin CE, Fields EC, McGuire WP, Temkin SM. Co-occurring substance use disorder: The impact on treatment adherence in women with locally advanced cervical cancer. Gynecol Oncol Rep. 2019;28:116-119. Published 2019 Mar 27. doi:10.1016/j.gore.2019.03.016
8. Chhatre S, Metzger DS, Malkowicz SB, Woody G, Jayadevappa R. Substance use disorder and its effects on outcomes in men with advanced-stage prostate cancer. Cancer. 2014;120(21):3338-3345. doi:10.1002/cncr.28861
9. Concannon K, Thayer JH, Hicks R, et al. Outcomes among patients with a history of substance abuse in non-small cell lung cancer: a county hospital experience. J Clin Onc. 2019;37(15)(suppl):e20031-e20031. doi:10.1200/JCO.2019.37.15
10. National Comprehensive Cancer Network. NCCN clinical practice guidelines in oncology: pancreatic adenocarcinoma. Version 2.2021. Updated February 25, 2021. Accessed July 8, 2021. https://www.nccn.org/professionals/physician_gls/pdf/pancreatic.pdf
11. Conroy T, Desseigne F, Ychou M, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011;364(19):1817-1825. doi:10.1056/NEJMoa1011923
12. Von Hoff DD, Ervin T, Arena FP, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med. 2013;369(18):1691-1703. doi:10.1056/NEJMoa1304369
13. Seal KH, Cohen G, Waldrop A, Cohen BE, Maguen S, Ren L. Substance use disorders in Iraq and Afghanistan veterans in VA healthcare, 2001-2010: Implications for screening, diagnosis and treatment. Drug Alcohol Depend. 2011;116(1-3):93-101. doi:10.1016/j.drugalcdep.2010.11.027
14. American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 5th ed. American Psychiatric Association; 2013.
A multidisciplinary approach provided safe and feasible cancer treatment in a patient with advanced pancreatic cancer and coexisting active substance use disorder.
A multidisciplinary approach provided safe and feasible cancer treatment in a patient with advanced pancreatic cancer and coexisting active substance use disorder.
Substance use disorders (SUDs) are an important but understudied aspect of treating patients diagnosed with cancer. Substance use can affect cancer treatment outcomes, including morbidity and mortality.1,2 Additionally, patients with cancer and SUD may have unique psychosocial needs that require close attention and management. There is a paucity of data regarding the best approach to treating such patients. For example, cocaine use may increase the cardiovascular and hematologic risk of some traditional chemotherapy agents.3,4 Newer targeted agents and immunotherapies remain understudied with respect to SUD risk.
Although the US Department of Veterans Affairs (VA) has established helpful clinical practice guidelines for the treatment of SUD, there are no guidelines for treating patients with SUD and cancer.5 Clinicians have limited confidence in treatment approach, and treatment is inconsistent among oncologists nationwide even within the same practice. Furthermore, it can be challenging to safely prescribe opioids for cancer-related pain in individuals with SUD. There is a high risk of SUD and mental health disorders in veterans, making this population particularly vulnerable. We report a case of a male with metastatic pancreatic cancer, severe opioid use disorder (OUD) and moderate cocaine use disorder (CUD) who received pain management and cancer treatment under the direction of a multidisciplinary team approach.
Case Report
A 63-year-old male with a medical history of HIV treated with highly active antiretroviral therapy (HAART), compensated cirrhosis, severe OUD, moderate CUD, and sedative use disorder in sustained remission was admitted to the West Haven campus of the VA Connecticut Healthcare System (VACHS) with abdominal pain, weight loss and fatigue. He used heroin 1 month prior to his admission and reported regular cocaine and marijuana use (Table 1). He was diagnosed with HIV in 1989, and his medical history included herpes zoster and oral candidiasis but no other opportunistic infections. Several months prior to this admission, he had an undetectable viral load and CD4 count of 688.
At the time of this admission, the patient was adherent to methadone treatment. He reported increased abdominal pain. Computed tomography (CT) showed a 2.4-cm mass in the pancreatic uncinate process, multiple liver metastases, retroperitoneal lymphadenopathy, and small lung nodules. A CT-guided liver biopsy showed adenocarcinoma consistent with a primary cancer of the pancreas. Given the complexity of the case, a multidisciplinary team approach was used to treat his cancer and the sequelae safely, including the oncology team, community living center team, palliative care team, and interprofessional opioid reassessment clinic team (ORC).
Cancer Treatment
Chemotherapy with FOLFIRINOX (leucovorin calcium, fluorouracil, irinotecan hydrochloride, and oxaliplatin) was recommended. The first cycle of treatment originally was planned for the outpatient setting, and a peripherally inserted central catheter (PICC) line was placed. However, after a urine toxicology test was positive for cocaine, the PICC line was removed due to concern for possible use of PICC line for nonprescribed substance use. The patient expressed suicidal ideation at the time and was admitted for psychiatric consult and pain control. Cycle 1 FOLFIRINOX was started during this admission. A PICC line was again put in place and then removed before discharge. A celiac plexus block was performed several days after this admission for pain control.
Given concern about cocaine use increasing the risk of cardiac toxicity with FOLFIRINOX treatment, treating providers sconsulted with the community living center (CLC) about possible admission for future chemotherapy administration and pain management. The CLC at VACHS has 38 beds for rehabilitation, long-term care, and hospice with the mission to restore each veteran to his or her highest level of well-being. After discussion with this patient and CLC staff, he agreed to a CLC admission. The patient agreed to remain in the facility, wear a secure care device, and not leave without staff accompaniment. He was able to obtain a 2-hour pass to pay bills and rent. During the 2 months he was admitted to the CLC he would present to the VACHS Cancer Center for chemotherapy every 2 weeks. He completed 6 cycles of chemotherapy while admitted. During the admission, he was transferred to active medical service for 2 days for fever and malaise, and then returned to the CLC. The patient elected to leave the CLC after 2 months as the inability to see close friends was interfering with his quality of life.
Upon being discharged from the CLC, shared decision making took place with the patient to establish a new treatment plan. In collaboration with the patient, a plan was made to admit him every 2 weeks for continued chemotherapy. A PICC line was placed on each day of admission and removed prior to discharge. It was also agreed that treatment would be delayed if a urine drug test was positive for cocaine on the morning of admission. The patient was also seen by ORC every 2 weeks after being discharged from the CLC.
Imaging after cycle 6 showed decreased size of liver metastases, retroperitoneal lymph nodes, and pancreas mass. Cancer antigen 19-9 (CA19-9) tumor marker was reduced from 3513 U/mL pretreatment to 50 U/mL after cycle 7. Chemotherapy cycle 7 was delayed 6 days due to active cocaine and heroin use. A repeat urine was obtained several days later, which was negative for cocaine, and he was admitted for cycle 7 chemotherapy. Using this treatment approach of admissions for every cycle, the patient was able to receive 11 cycles of FOLFIRINOX with clinical benefit.
Palliative Care/Pain Management
Safely treating the patient’s malignant pain in the context of his OUD was critically important. In order to do this the palliative care team worked closely alongside ORC, is a multidisciplinary team consisting of health care providers (HCPs) from addiction psychiatry, internal medicine, health psychology and pharmacy who are consulted to evaluate veterans’ current opioid regimens and make recommendations to optimize both safety and efficacy. ORC followed this particular veteran as an outpatient and consulted on pain issues during his admission. They recommended the continuation of methadone at 120 mg daily and increased oral oxycodone to 30 mg every 6 hours, and then further increased to 45 mg every 6 hours. He continued to have increased pain despite higher doses of oxycodone, and pain medication was changed to oral hydromorphone 28 mg every 6 hours with the continuation of methadone. ORC and the palliative care team obtained consent from the veteran and a release of Information form signed by the patient to contact his community methadone clinic for further collaboration around pain management throughout the time caring for the veteran.
Even with improvement in disease based on imaging and tumor markers, opioid medications could not be decreased in this case. This is likely in part due to the multidimensional nature of pain. Careful assessment of the biologic, emotional, social, and spiritual contributors to pain is needed in the management of pain, especially at end of life.6 Nonpharmacologic pain management strategies used in this case included a transcutaneous electrical nerve stimulation unit, moist heat, celiac plexus block, and emotional support.
Psychosocial Issues/Substance Use
Psychosocial support for the patient was provided by the interdisciplinary palliative care team and the ORC team in both the inpatient and outpatient settings. Despite efforts from case management to get the veteran home services once discharged from the CLC, he declined repeatedly. Thus, the CLC social worker obtained a guardian alert for the veteran on discharge.
Close outpatient follow-up for medical and psychosocial support was very critical. When an outpatient, the veteran was scheduled for biweekly appointments with palliative care or ORC. When admitted to the hospital, the palliative care team medical director and psychologist conducted joint visits with him. Although he denied depressed mood and anxiety throughout his treatment, he often reflected on regrets that he had as he faced the end of his life. Specifically, he shared thoughts about being estranged from his surviving brother given his long struggle with substance use. Although he did not think a relationship was possible with his brother at the end of life, he still cared deeply for him and wanted to make him aware of his pancreatic cancer diagnosis. This was particularly important to him because their late brother had also died of pancreatic cancer. It was the patient’s wish at the end of his life to alert his surviving brother of his diagnosis so he and his children could get adequate screening throughout their lives. Although he had spoken of this desire often, it wasn’t until his disease progressed and he elected to transition to hospice that he felt ready to write the letter. The palliative care team assisted the veteran in writing and mailing a letter to his brother informing him of his diagnosis and transition to hospice as well as communicating that his brother and his family had been in his thoughts at the end of his life. The patient’s brother received this letter and with assistance from the CLC social worker made arrangements to visit the veteran at bedside at the inpatient CLC hospice unit the final days of his life.
Discussion
There are very little data on the safety of cancer-directed therapy in patients with active SUD. The limited studies that have been done showed conflicting results.
A retrospective study among women with co-occurring SUD and locally advanced cervical cancer who were undergoing primary radiation therapy found that SUD was not associated with a difference in toxicity or survival outcomes.7 However, other research suggests that SUD may be associated with an increase in all-cause mortality as well as other adverse outcomes for patients and health care systems (eg, emergency department visits, hospitalizations).8 A retrospective study of patients with a history of SUD and nonsmall cell lung cancer showed that these patients had higher rates of depression, less family support, increased rates of missed appointments, more emergency department visits and more hospitalizations.9 Patients with chronic myeloid leukemia or myelodysplastic syndromes who had long-term cocaine use had a 6-fold increased risk of death, which was not found in patients who had long-term alcohol or marijuana use.2
The limited data highlight the need for careful consideration of ways to mitigate potentially adverse outcomes in this population while still providing clinically indicated cancer treatment. Integrated VA health care systems provide unique resources that can maximize veteran safety during cancer treatment. Utilization of VA resources and close interdisciplinary collaboration across VA HCPs can help to ensure equitable access to state-of-the-art cancer therapies for veterans with comorbid SUD.
VA Services for Patients With Comorbidities
This case highlights several distinct aspects of VA health care that make it possible to safely treat individuals with complex comorbidities. One important aspect of this was collaboration with the CLC to admit the veteran for his initial treatment after a positive cocaine test. CLC admission was nonpunitive and allowed ongoing involvement in the VA community. This provided an essential, safe, and structured environment in which 6 cycles of chemotherapy could be delivered.
Although the patient left the CLC after 2 months due to floor restrictions negatively impacting his quality of life and ability to spend time with close friends, several important events occurred during this stay. First, the patient established close relationships with the CLC staff and the palliative care team; both groups followed him throughout his inpatient and outpatient care. These relationships proved essential throughout his care as they were the foundation of difficult conversations about substance use, treatment adherence, and eventually, transition to hospice.
In addition, the opportunity to administer 6 cycles of chemotherapy at the CLC was enough to lead to clinical benefit and radiographic response to treatment. Clinical benefits while in the CLC included maintenance of a good appetite, 15-lb weight gain and preserved performance status (ECOG [Eastern Cooperative Group]-1), which allowed him to actively participate in multiple social and recreational activities while in the CLC. From early conversations, this patient was clear that he wanted treatment as long as his life could be prolonged with good quality of life. Having evidence of the benefit of treatment, at least initially, increased the patient’s confidence in treatment. There were a few conversations when the challenges of treatment mounted (eg, pain, needs for abstinence from cocaine prior to admission for chemotherapy, frequent doctor appointments), and the patient would remind himself of these data to recommit himself to treatment. The opportunity to admit him to the inpatient VA facility, including bed availability for 3 days during his treatment once he left the CLC was important. This plan to admit the patient following a negative urine toxicology test for cocaine was made collaboratively with the veteran and the oncology and palliative care teams. The plan allowed the patient to achieve his treatment goals while maintaining his safety and reducing theoretical cardiac toxicities with his cancer treatment.
Finally, the availability of a multidisciplinary team approach including palliative care, oncology, psychology, addiction medicine and addiction psychiatry, was critical for addressing the veteran’s malignant pain. Palliative care worked in close collaboration with the ORC to prescribe and renew pain medications. ORC offered ongoing consultation on pain management in the context of OUD. As the veteran’s cancer progressed and functional decline prohibited his daily attendance at the community methadone clinic, palliative care and ORC met with the methadone clinic to arrange a less frequent methadone pickup schedule (the patient previously needed daily pickup). Non-VA settings may not have access to these resources to safely treat the biopsychosocial issues that arise in complex cases.
Substance Use and Cancer Treatments
This case raises several critical questions for oncologic care. Cocaine and fluorouracil are both associated with cardiotoxicity, and many oncologists would not feel it is safe to administer a regimen containing fluorouracil to a patient with active cocaine use. The National Comprehensive Cancer Network (NCCN) panel recommends FOLFIRINOX as a preferred category 1 recommendation for first-line treatment of patients with advanced pancreas cancer with good performance status.10 This recommendation is based on the PRODIGE trial, which has shown improved overall survival (OS): 11.1 vs 6.8 months for patients who received single-agent gemcitabine.11 If patients are not candidates for FOLFIRINOX and have good performance status, the NCCN recommends gemcitabine plus albumin-bound paclitaxel with category 1 level of evidence based on the IMPACT trial, which showed improvement in OS (8.7 vs 6.6 months compared with single-agent gemcitabine).12
Some oncologists may have additional concerns administering fluorouracil treatment alternatives (such as gemcitabine and albumin-bound paclitaxel) to individuals with active SUD because of concerns about altered mental status impacting the ability to report important adverse effects. In the absence of sufficient data, HCPs must determine whether they feel it is safe to administer these agents in individuals with active cocaine use. However, denying these patients the possible benefits of standard-of-care life-prolonging therapies without established data raises concerns regarding the ethics of such practices. There is concern that the stigma surrounding cocaine use might contribute to withholding treatment, while treatment is continued for individuals taking prescribed stimulant medications that also have cardiotoxicity risks. VA health care facilities are uniquely situated to use all available resources to address these issues using interprofessional patient-centered care and determine the most optimal treatment based on a risk/benefit discussion between the patient and the HCP.
Similarly, this case also raised questions among HCPs about the safety of using an indwelling port for treatment in a patient with SUD. In the current case there was concern about keeping in a port for a patient with a history of IV drug use; therefore, a PICC line was initiated and removed at each admission. Without guidelines in these situations, HCPs are left to weigh the risks and benefits of using a port or a PICC for individuals with recent or current substance use without formal data, which can lead to inconsistent access to care. More guidance is needed for these situations.
SUD Screening
This case begs the question of whether oncologists are adequately screening for a range of SUDs, and when they encounter an issue, how they are addressing it. Many oncologists do not receive adequate training on assessment of current or recent substance use. There are health care and systems-level practices that may increase patient safety for individuals with ongoing substance use who are undergoing cancer treatment. Training on obtaining appropriate substance use histories, motivational interviewing to resolve ambivalence about substance use in the direction of change, and shared decision making about treatment options could increase confidence in understanding and addressing substance use issues. It is also important to educate oncologists on how to address patients who return to or continued substance use during treatment. In this case the collaboration from palliative care, psychology, addiction medicine, and addiction psychiatry through the ORC was essential in assisting with ongoing assessment of substance use, guiding difficult conversations about the impact of substance use on the treatment plan, and identifying risk-mitigation strategies. Close collaboration and full utilization of all VA resources allowed this patient to receive first-line treatment for pancreatic cancer in order to reach his goal of prolonging his life while maintaining acceptable quality of life. Table 2 provides best practices for management of patients with comorbid SUD and cancer.
More research is needed into cancer treatment for patients with SUD, especially in the current era of cancer care using novel cancer treatments leading to significantly improved survival in many cancer types. Ideally, oncologists should be routinely or consistently screening patients for substance use, including alcohol. The patient should participate in this decision-making process after being educated about the risks and benefits. These patients can be followed using a multimodal approach to increase their rates of success and improve their quality of life. Although the literature is limited and no formal guidelines are available, VA oncologists are fortunate to have a range of resources available to them to navigate these difficult cases. Veterans have elevated rates of SUD, making this a critical issue to consider in the VA.13 It is the hope that this case can highlight how to take advantage of the many VA resources in order to ensure equitable cancer care for all veterans.
Conclusions
This case demonstrates that cancer-directed treatment is safe and feasible in a patient with advanced pancreatic cancer and coexisting active SUD by using a multidisciplinary approach. The multidisciplinary team included palliative care, oncology, psychology, addiction medicine, and addiction psychiatry. Critical steps for a successful outcome include gathering history about SUD; motivational interviewing to resolve ambivalence about treatment for SUD; shared decision making about cancer treatment; and risk-reduction strategies in pain and SUD management.
Treatment advancements in many cancer types have led to significantly longer survival, and it is critical to develop safe protocols to treat patients with active SUD so they also can derive benefit from these very significant medical advancements.
Acknowledgments
Michal Rose, MD, Director of VACHS Cancer Center, and Chandrika Kumar, MD, Director of VACHS Community Living Center, for their collaboration in care for this veteran.
Substance use disorders (SUDs) are an important but understudied aspect of treating patients diagnosed with cancer. Substance use can affect cancer treatment outcomes, including morbidity and mortality.1,2 Additionally, patients with cancer and SUD may have unique psychosocial needs that require close attention and management. There is a paucity of data regarding the best approach to treating such patients. For example, cocaine use may increase the cardiovascular and hematologic risk of some traditional chemotherapy agents.3,4 Newer targeted agents and immunotherapies remain understudied with respect to SUD risk.
Although the US Department of Veterans Affairs (VA) has established helpful clinical practice guidelines for the treatment of SUD, there are no guidelines for treating patients with SUD and cancer.5 Clinicians have limited confidence in treatment approach, and treatment is inconsistent among oncologists nationwide even within the same practice. Furthermore, it can be challenging to safely prescribe opioids for cancer-related pain in individuals with SUD. There is a high risk of SUD and mental health disorders in veterans, making this population particularly vulnerable. We report a case of a male with metastatic pancreatic cancer, severe opioid use disorder (OUD) and moderate cocaine use disorder (CUD) who received pain management and cancer treatment under the direction of a multidisciplinary team approach.
Case Report
A 63-year-old male with a medical history of HIV treated with highly active antiretroviral therapy (HAART), compensated cirrhosis, severe OUD, moderate CUD, and sedative use disorder in sustained remission was admitted to the West Haven campus of the VA Connecticut Healthcare System (VACHS) with abdominal pain, weight loss and fatigue. He used heroin 1 month prior to his admission and reported regular cocaine and marijuana use (Table 1). He was diagnosed with HIV in 1989, and his medical history included herpes zoster and oral candidiasis but no other opportunistic infections. Several months prior to this admission, he had an undetectable viral load and CD4 count of 688.
At the time of this admission, the patient was adherent to methadone treatment. He reported increased abdominal pain. Computed tomography (CT) showed a 2.4-cm mass in the pancreatic uncinate process, multiple liver metastases, retroperitoneal lymphadenopathy, and small lung nodules. A CT-guided liver biopsy showed adenocarcinoma consistent with a primary cancer of the pancreas. Given the complexity of the case, a multidisciplinary team approach was used to treat his cancer and the sequelae safely, including the oncology team, community living center team, palliative care team, and interprofessional opioid reassessment clinic team (ORC).
Cancer Treatment
Chemotherapy with FOLFIRINOX (leucovorin calcium, fluorouracil, irinotecan hydrochloride, and oxaliplatin) was recommended. The first cycle of treatment originally was planned for the outpatient setting, and a peripherally inserted central catheter (PICC) line was placed. However, after a urine toxicology test was positive for cocaine, the PICC line was removed due to concern for possible use of PICC line for nonprescribed substance use. The patient expressed suicidal ideation at the time and was admitted for psychiatric consult and pain control. Cycle 1 FOLFIRINOX was started during this admission. A PICC line was again put in place and then removed before discharge. A celiac plexus block was performed several days after this admission for pain control.
Given concern about cocaine use increasing the risk of cardiac toxicity with FOLFIRINOX treatment, treating providers sconsulted with the community living center (CLC) about possible admission for future chemotherapy administration and pain management. The CLC at VACHS has 38 beds for rehabilitation, long-term care, and hospice with the mission to restore each veteran to his or her highest level of well-being. After discussion with this patient and CLC staff, he agreed to a CLC admission. The patient agreed to remain in the facility, wear a secure care device, and not leave without staff accompaniment. He was able to obtain a 2-hour pass to pay bills and rent. During the 2 months he was admitted to the CLC he would present to the VACHS Cancer Center for chemotherapy every 2 weeks. He completed 6 cycles of chemotherapy while admitted. During the admission, he was transferred to active medical service for 2 days for fever and malaise, and then returned to the CLC. The patient elected to leave the CLC after 2 months as the inability to see close friends was interfering with his quality of life.
Upon being discharged from the CLC, shared decision making took place with the patient to establish a new treatment plan. In collaboration with the patient, a plan was made to admit him every 2 weeks for continued chemotherapy. A PICC line was placed on each day of admission and removed prior to discharge. It was also agreed that treatment would be delayed if a urine drug test was positive for cocaine on the morning of admission. The patient was also seen by ORC every 2 weeks after being discharged from the CLC.
Imaging after cycle 6 showed decreased size of liver metastases, retroperitoneal lymph nodes, and pancreas mass. Cancer antigen 19-9 (CA19-9) tumor marker was reduced from 3513 U/mL pretreatment to 50 U/mL after cycle 7. Chemotherapy cycle 7 was delayed 6 days due to active cocaine and heroin use. A repeat urine was obtained several days later, which was negative for cocaine, and he was admitted for cycle 7 chemotherapy. Using this treatment approach of admissions for every cycle, the patient was able to receive 11 cycles of FOLFIRINOX with clinical benefit.
Palliative Care/Pain Management
Safely treating the patient’s malignant pain in the context of his OUD was critically important. In order to do this the palliative care team worked closely alongside ORC, is a multidisciplinary team consisting of health care providers (HCPs) from addiction psychiatry, internal medicine, health psychology and pharmacy who are consulted to evaluate veterans’ current opioid regimens and make recommendations to optimize both safety and efficacy. ORC followed this particular veteran as an outpatient and consulted on pain issues during his admission. They recommended the continuation of methadone at 120 mg daily and increased oral oxycodone to 30 mg every 6 hours, and then further increased to 45 mg every 6 hours. He continued to have increased pain despite higher doses of oxycodone, and pain medication was changed to oral hydromorphone 28 mg every 6 hours with the continuation of methadone. ORC and the palliative care team obtained consent from the veteran and a release of Information form signed by the patient to contact his community methadone clinic for further collaboration around pain management throughout the time caring for the veteran.
Even with improvement in disease based on imaging and tumor markers, opioid medications could not be decreased in this case. This is likely in part due to the multidimensional nature of pain. Careful assessment of the biologic, emotional, social, and spiritual contributors to pain is needed in the management of pain, especially at end of life.6 Nonpharmacologic pain management strategies used in this case included a transcutaneous electrical nerve stimulation unit, moist heat, celiac plexus block, and emotional support.
Psychosocial Issues/Substance Use
Psychosocial support for the patient was provided by the interdisciplinary palliative care team and the ORC team in both the inpatient and outpatient settings. Despite efforts from case management to get the veteran home services once discharged from the CLC, he declined repeatedly. Thus, the CLC social worker obtained a guardian alert for the veteran on discharge.
Close outpatient follow-up for medical and psychosocial support was very critical. When an outpatient, the veteran was scheduled for biweekly appointments with palliative care or ORC. When admitted to the hospital, the palliative care team medical director and psychologist conducted joint visits with him. Although he denied depressed mood and anxiety throughout his treatment, he often reflected on regrets that he had as he faced the end of his life. Specifically, he shared thoughts about being estranged from his surviving brother given his long struggle with substance use. Although he did not think a relationship was possible with his brother at the end of life, he still cared deeply for him and wanted to make him aware of his pancreatic cancer diagnosis. This was particularly important to him because their late brother had also died of pancreatic cancer. It was the patient’s wish at the end of his life to alert his surviving brother of his diagnosis so he and his children could get adequate screening throughout their lives. Although he had spoken of this desire often, it wasn’t until his disease progressed and he elected to transition to hospice that he felt ready to write the letter. The palliative care team assisted the veteran in writing and mailing a letter to his brother informing him of his diagnosis and transition to hospice as well as communicating that his brother and his family had been in his thoughts at the end of his life. The patient’s brother received this letter and with assistance from the CLC social worker made arrangements to visit the veteran at bedside at the inpatient CLC hospice unit the final days of his life.
Discussion
There are very little data on the safety of cancer-directed therapy in patients with active SUD. The limited studies that have been done showed conflicting results.
A retrospective study among women with co-occurring SUD and locally advanced cervical cancer who were undergoing primary radiation therapy found that SUD was not associated with a difference in toxicity or survival outcomes.7 However, other research suggests that SUD may be associated with an increase in all-cause mortality as well as other adverse outcomes for patients and health care systems (eg, emergency department visits, hospitalizations).8 A retrospective study of patients with a history of SUD and nonsmall cell lung cancer showed that these patients had higher rates of depression, less family support, increased rates of missed appointments, more emergency department visits and more hospitalizations.9 Patients with chronic myeloid leukemia or myelodysplastic syndromes who had long-term cocaine use had a 6-fold increased risk of death, which was not found in patients who had long-term alcohol or marijuana use.2
The limited data highlight the need for careful consideration of ways to mitigate potentially adverse outcomes in this population while still providing clinically indicated cancer treatment. Integrated VA health care systems provide unique resources that can maximize veteran safety during cancer treatment. Utilization of VA resources and close interdisciplinary collaboration across VA HCPs can help to ensure equitable access to state-of-the-art cancer therapies for veterans with comorbid SUD.
VA Services for Patients With Comorbidities
This case highlights several distinct aspects of VA health care that make it possible to safely treat individuals with complex comorbidities. One important aspect of this was collaboration with the CLC to admit the veteran for his initial treatment after a positive cocaine test. CLC admission was nonpunitive and allowed ongoing involvement in the VA community. This provided an essential, safe, and structured environment in which 6 cycles of chemotherapy could be delivered.
Although the patient left the CLC after 2 months due to floor restrictions negatively impacting his quality of life and ability to spend time with close friends, several important events occurred during this stay. First, the patient established close relationships with the CLC staff and the palliative care team; both groups followed him throughout his inpatient and outpatient care. These relationships proved essential throughout his care as they were the foundation of difficult conversations about substance use, treatment adherence, and eventually, transition to hospice.
In addition, the opportunity to administer 6 cycles of chemotherapy at the CLC was enough to lead to clinical benefit and radiographic response to treatment. Clinical benefits while in the CLC included maintenance of a good appetite, 15-lb weight gain and preserved performance status (ECOG [Eastern Cooperative Group]-1), which allowed him to actively participate in multiple social and recreational activities while in the CLC. From early conversations, this patient was clear that he wanted treatment as long as his life could be prolonged with good quality of life. Having evidence of the benefit of treatment, at least initially, increased the patient’s confidence in treatment. There were a few conversations when the challenges of treatment mounted (eg, pain, needs for abstinence from cocaine prior to admission for chemotherapy, frequent doctor appointments), and the patient would remind himself of these data to recommit himself to treatment. The opportunity to admit him to the inpatient VA facility, including bed availability for 3 days during his treatment once he left the CLC was important. This plan to admit the patient following a negative urine toxicology test for cocaine was made collaboratively with the veteran and the oncology and palliative care teams. The plan allowed the patient to achieve his treatment goals while maintaining his safety and reducing theoretical cardiac toxicities with his cancer treatment.
Finally, the availability of a multidisciplinary team approach including palliative care, oncology, psychology, addiction medicine and addiction psychiatry, was critical for addressing the veteran’s malignant pain. Palliative care worked in close collaboration with the ORC to prescribe and renew pain medications. ORC offered ongoing consultation on pain management in the context of OUD. As the veteran’s cancer progressed and functional decline prohibited his daily attendance at the community methadone clinic, palliative care and ORC met with the methadone clinic to arrange a less frequent methadone pickup schedule (the patient previously needed daily pickup). Non-VA settings may not have access to these resources to safely treat the biopsychosocial issues that arise in complex cases.
Substance Use and Cancer Treatments
This case raises several critical questions for oncologic care. Cocaine and fluorouracil are both associated with cardiotoxicity, and many oncologists would not feel it is safe to administer a regimen containing fluorouracil to a patient with active cocaine use. The National Comprehensive Cancer Network (NCCN) panel recommends FOLFIRINOX as a preferred category 1 recommendation for first-line treatment of patients with advanced pancreas cancer with good performance status.10 This recommendation is based on the PRODIGE trial, which has shown improved overall survival (OS): 11.1 vs 6.8 months for patients who received single-agent gemcitabine.11 If patients are not candidates for FOLFIRINOX and have good performance status, the NCCN recommends gemcitabine plus albumin-bound paclitaxel with category 1 level of evidence based on the IMPACT trial, which showed improvement in OS (8.7 vs 6.6 months compared with single-agent gemcitabine).12
Some oncologists may have additional concerns administering fluorouracil treatment alternatives (such as gemcitabine and albumin-bound paclitaxel) to individuals with active SUD because of concerns about altered mental status impacting the ability to report important adverse effects. In the absence of sufficient data, HCPs must determine whether they feel it is safe to administer these agents in individuals with active cocaine use. However, denying these patients the possible benefits of standard-of-care life-prolonging therapies without established data raises concerns regarding the ethics of such practices. There is concern that the stigma surrounding cocaine use might contribute to withholding treatment, while treatment is continued for individuals taking prescribed stimulant medications that also have cardiotoxicity risks. VA health care facilities are uniquely situated to use all available resources to address these issues using interprofessional patient-centered care and determine the most optimal treatment based on a risk/benefit discussion between the patient and the HCP.
Similarly, this case also raised questions among HCPs about the safety of using an indwelling port for treatment in a patient with SUD. In the current case there was concern about keeping in a port for a patient with a history of IV drug use; therefore, a PICC line was initiated and removed at each admission. Without guidelines in these situations, HCPs are left to weigh the risks and benefits of using a port or a PICC for individuals with recent or current substance use without formal data, which can lead to inconsistent access to care. More guidance is needed for these situations.
SUD Screening
This case begs the question of whether oncologists are adequately screening for a range of SUDs, and when they encounter an issue, how they are addressing it. Many oncologists do not receive adequate training on assessment of current or recent substance use. There are health care and systems-level practices that may increase patient safety for individuals with ongoing substance use who are undergoing cancer treatment. Training on obtaining appropriate substance use histories, motivational interviewing to resolve ambivalence about substance use in the direction of change, and shared decision making about treatment options could increase confidence in understanding and addressing substance use issues. It is also important to educate oncologists on how to address patients who return to or continued substance use during treatment. In this case the collaboration from palliative care, psychology, addiction medicine, and addiction psychiatry through the ORC was essential in assisting with ongoing assessment of substance use, guiding difficult conversations about the impact of substance use on the treatment plan, and identifying risk-mitigation strategies. Close collaboration and full utilization of all VA resources allowed this patient to receive first-line treatment for pancreatic cancer in order to reach his goal of prolonging his life while maintaining acceptable quality of life. Table 2 provides best practices for management of patients with comorbid SUD and cancer.
More research is needed into cancer treatment for patients with SUD, especially in the current era of cancer care using novel cancer treatments leading to significantly improved survival in many cancer types. Ideally, oncologists should be routinely or consistently screening patients for substance use, including alcohol. The patient should participate in this decision-making process after being educated about the risks and benefits. These patients can be followed using a multimodal approach to increase their rates of success and improve their quality of life. Although the literature is limited and no formal guidelines are available, VA oncologists are fortunate to have a range of resources available to them to navigate these difficult cases. Veterans have elevated rates of SUD, making this a critical issue to consider in the VA.13 It is the hope that this case can highlight how to take advantage of the many VA resources in order to ensure equitable cancer care for all veterans.
Conclusions
This case demonstrates that cancer-directed treatment is safe and feasible in a patient with advanced pancreatic cancer and coexisting active SUD by using a multidisciplinary approach. The multidisciplinary team included palliative care, oncology, psychology, addiction medicine, and addiction psychiatry. Critical steps for a successful outcome include gathering history about SUD; motivational interviewing to resolve ambivalence about treatment for SUD; shared decision making about cancer treatment; and risk-reduction strategies in pain and SUD management.
Treatment advancements in many cancer types have led to significantly longer survival, and it is critical to develop safe protocols to treat patients with active SUD so they also can derive benefit from these very significant medical advancements.
Acknowledgments
Michal Rose, MD, Director of VACHS Cancer Center, and Chandrika Kumar, MD, Director of VACHS Community Living Center, for their collaboration in care for this veteran.
1. Chang G, Meadows ME, Jones JA, Antin JH, Orav EJ. Substance use and survival after treatment for chronic myelogenous leukemia (CML) or myelodysplastic syndrome (MDS). Am J Drug Alcohol Ab. 2010;36(1):1-6. doi:10.3109/00952990903490758
2. Stagno S, Busby K, Shapiro A, Kotz M. Patients at risk: addressing addiction in patients undergoing hematopoietic SCT. Bone Marrow Transplant. 2008;42(4):221-226. doi:10.1038/bmt.2008.211
3. Arora NP. Cutaneous vasculopathy and neutropenia associated with levamisole-adulterated cocaine. Am J Med Sci. 2013;345(1):45-51. doi:10.1097/MAJ.0b013e31825b2b50
4. Schwartz BG, Rezkalla S, Kloner RA. Cardiovascular effects of cocaine. Circulation. 2010;122(24):2558-2569. doi:10.1161/CIRCULATIONAHA.110.940569
5. US Department of Veterans Affairs, US Department of Defense. VA/DoD clinical practice guideline for the management of substance use disorders. Published 2015. Accessed July 8, 2021. https://www.healthquality.va.gov/guidelines/MH/sud/VADODSUDCPGRevised22216.pdf
6. Mehta A, Chan LS. Understanding of the concept of “total pain”: a prerequisite for pain control. J Hosp Palliat Nurs. 2008;10(1):26-32. doi:10.1097/01.NJH.0000306714.50539.1a
7. Rubinsak LA, Terplan M, Martin CE, Fields EC, McGuire WP, Temkin SM. Co-occurring substance use disorder: The impact on treatment adherence in women with locally advanced cervical cancer. Gynecol Oncol Rep. 2019;28:116-119. Published 2019 Mar 27. doi:10.1016/j.gore.2019.03.016
8. Chhatre S, Metzger DS, Malkowicz SB, Woody G, Jayadevappa R. Substance use disorder and its effects on outcomes in men with advanced-stage prostate cancer. Cancer. 2014;120(21):3338-3345. doi:10.1002/cncr.28861
9. Concannon K, Thayer JH, Hicks R, et al. Outcomes among patients with a history of substance abuse in non-small cell lung cancer: a county hospital experience. J Clin Onc. 2019;37(15)(suppl):e20031-e20031. doi:10.1200/JCO.2019.37.15
10. National Comprehensive Cancer Network. NCCN clinical practice guidelines in oncology: pancreatic adenocarcinoma. Version 2.2021. Updated February 25, 2021. Accessed July 8, 2021. https://www.nccn.org/professionals/physician_gls/pdf/pancreatic.pdf
11. Conroy T, Desseigne F, Ychou M, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011;364(19):1817-1825. doi:10.1056/NEJMoa1011923
12. Von Hoff DD, Ervin T, Arena FP, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med. 2013;369(18):1691-1703. doi:10.1056/NEJMoa1304369
13. Seal KH, Cohen G, Waldrop A, Cohen BE, Maguen S, Ren L. Substance use disorders in Iraq and Afghanistan veterans in VA healthcare, 2001-2010: Implications for screening, diagnosis and treatment. Drug Alcohol Depend. 2011;116(1-3):93-101. doi:10.1016/j.drugalcdep.2010.11.027
14. American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 5th ed. American Psychiatric Association; 2013.
1. Chang G, Meadows ME, Jones JA, Antin JH, Orav EJ. Substance use and survival after treatment for chronic myelogenous leukemia (CML) or myelodysplastic syndrome (MDS). Am J Drug Alcohol Ab. 2010;36(1):1-6. doi:10.3109/00952990903490758
2. Stagno S, Busby K, Shapiro A, Kotz M. Patients at risk: addressing addiction in patients undergoing hematopoietic SCT. Bone Marrow Transplant. 2008;42(4):221-226. doi:10.1038/bmt.2008.211
3. Arora NP. Cutaneous vasculopathy and neutropenia associated with levamisole-adulterated cocaine. Am J Med Sci. 2013;345(1):45-51. doi:10.1097/MAJ.0b013e31825b2b50
4. Schwartz BG, Rezkalla S, Kloner RA. Cardiovascular effects of cocaine. Circulation. 2010;122(24):2558-2569. doi:10.1161/CIRCULATIONAHA.110.940569
5. US Department of Veterans Affairs, US Department of Defense. VA/DoD clinical practice guideline for the management of substance use disorders. Published 2015. Accessed July 8, 2021. https://www.healthquality.va.gov/guidelines/MH/sud/VADODSUDCPGRevised22216.pdf
6. Mehta A, Chan LS. Understanding of the concept of “total pain”: a prerequisite for pain control. J Hosp Palliat Nurs. 2008;10(1):26-32. doi:10.1097/01.NJH.0000306714.50539.1a
7. Rubinsak LA, Terplan M, Martin CE, Fields EC, McGuire WP, Temkin SM. Co-occurring substance use disorder: The impact on treatment adherence in women with locally advanced cervical cancer. Gynecol Oncol Rep. 2019;28:116-119. Published 2019 Mar 27. doi:10.1016/j.gore.2019.03.016
8. Chhatre S, Metzger DS, Malkowicz SB, Woody G, Jayadevappa R. Substance use disorder and its effects on outcomes in men with advanced-stage prostate cancer. Cancer. 2014;120(21):3338-3345. doi:10.1002/cncr.28861
9. Concannon K, Thayer JH, Hicks R, et al. Outcomes among patients with a history of substance abuse in non-small cell lung cancer: a county hospital experience. J Clin Onc. 2019;37(15)(suppl):e20031-e20031. doi:10.1200/JCO.2019.37.15
10. National Comprehensive Cancer Network. NCCN clinical practice guidelines in oncology: pancreatic adenocarcinoma. Version 2.2021. Updated February 25, 2021. Accessed July 8, 2021. https://www.nccn.org/professionals/physician_gls/pdf/pancreatic.pdf
11. Conroy T, Desseigne F, Ychou M, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011;364(19):1817-1825. doi:10.1056/NEJMoa1011923
12. Von Hoff DD, Ervin T, Arena FP, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med. 2013;369(18):1691-1703. doi:10.1056/NEJMoa1304369
13. Seal KH, Cohen G, Waldrop A, Cohen BE, Maguen S, Ren L. Substance use disorders in Iraq and Afghanistan veterans in VA healthcare, 2001-2010: Implications for screening, diagnosis and treatment. Drug Alcohol Depend. 2011;116(1-3):93-101. doi:10.1016/j.drugalcdep.2010.11.027
14. American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 5th ed. American Psychiatric Association; 2013.
Constipation and Postprandial Pain in a Patient With Shortness of Breath
A 62-year-old male veteran with a history of pulmonary embolism (PE) and prostate cancer status after brachytherapy presented to the emergency department with new onset shortness of breath and left-sided chest pain after prolonged car travel. He underwent a chest computed tomography (CT) angiogram that showed no PE recurrence; however, the scan revealed an incidental transverse colon mass that appeared well circumscribed, homogeneous, and radiolucent with no enhancement, septations, or hypervascularity but no evidence of colonic distension or obstruction (Figure 1).
- What is your diagnosis?
- How would you treat this patient?
The patient reported having chronic constipation and a dull, left-sided abdominal discomfort for the past year. He noted that his abdominal pain worsened after eating and mildly improved after taking castor oil. He had no surgical history and no family history of cancer. The patient reported no fever, fatigue, weight loss, chills, nausea, vomiting, diarrhea, hematochezia, dysuria, hematuria, or melena. Vital signs, physical examination, and initial routine laboratory work were all within appropriate ranges, and a fecal occult blood test was negative.
A colonoscopy was performed, revealing a near-obstructing submucosal mass in the transverse colon near the splenic flexure with a smooth surface and a positive Cushion (Pillow) sign (Figure 2). The patient underwent surgical exploration that resulted in finding a soft, 11-cm lesion arising from the mesenteric side of the transverse colon (Figure 3). Hematoxylin and eosin (H&E) stains were used on a sample from the mass (Figure 4).
The tumor was enucleated via a colotomy over the mass, and the colotomy repaired primarily. Gross examination revealed homogenous yellow fatty tissue, and the H&E stains showed mature, well-differentiated adipocytes with uniform nuclei surrounded by a fibrous capsule. Based on this pathologic examination, this patient was diagnosed with a lipoma of the transverse colon. The resected tissue showed negative margins, indicating full removal of the lipoma.
The patient stabilized well after surgery and remained under inpatient care for observation; due to lack of appetite following the surgery, the patient did not start eating solids again until 2 days after the lipoma removal. By postoperative day 4, the patient had return of bowel function and was tolerating a regular diet with no recurrence of his prandial pain, shortness of breath, or left-sided chest pain. While the precise cause of the patient’s initial presentation of shortness of breath and left-sided chest pain was not ascertained, it is likely that the lipoma, near completely obstructed his bowel, caused abdominal contents and distended intestines to push against his diaphragm, leading to pain and dyspnea. This was likely exacerbated by sensitization to these symptoms from his prior PE. He was discharged home on postoperative day 4 with outpatient follow-up with general surgery.
Discussion
Lipomas are common benign tumors arising from aberrantly multiplying adipocytes. Although lipomas are most commonly found subcutaneously, the lesions can occur anywhere along the gastrointestinal (GI) tract, most often in the colon.1 The incidence rate of colon lipomas ranges from 0.2 to 4.4% among patients in their fifth to sixth decades of life, more commonly found in females.2 These lesions are the most common submucosal mesenchymal lesions of the colon, with a predilection for the right ascending colon.1 The etiology of colon lipomas is largely unknown; one known cause is trauma, thought to induce cytokine release or HMGA2-LPP fusion gene arrangements leading to adipocyte proliferation.3
Most colon lipomas are asymptomatic and discovered incidentally; symptoms typically arise when the lesions are > 2 cm in diameter and include abdominal pain, changes in bowel habits, rectal bleeding, and in extreme cases, obstruction and perforation.4 On CT imaging, colon lipomas will appear radiolucent, homogenous, and well circumscribed. The lesions usually do not warrant intervention unless they are symptomatic. If symptomatic, resection of the lesion is the first-line treatment and usually results in complete resolution of symptoms with no recurrence.2
While either a surgical or endoscopic approach may be used for resection, an increased risk of perforation of the colon with larger lipomas has been shown with endoscopic excision.5 With surgical resection, an open or minimally invasive approach may be offered, based on surgeon comfort with minimally invasive colon procedures. Minimally invasive colonic surgeries may be associated with a shorter length of stay, decreased postoperative pain, and faster return of bowel function. In this case, the surgeon chose an open approach due to the large size of the mass (11 cm) as well as location of the mass in the transverse colon, which made it easy to access directly through a small laparotomy incision made in the superior midline over the transverse colon.
When a colonic mesenchymal mass is seen on colonoscopy, it is important to consider other, nonbenign lesions that present this way. The most common malignant mesenchymal tumor of the GI tract is a gastrointestinal stromal tumor (GIST), a soft-tissue sarcoma that occurs predominantly in the stomach and small intestine.6 These tumors arise from the interstitial cells of Cajal (ICC) and are associated with mutations of KIT and PDGFR-α genes.7 The incidence in the United States is approximately 0.70 per 100,000 people per year, predominantly found in adults in their fifth or sixth decade of life.8 While this tumor typically occurs in the upper GI tract, very rarely, GISTs can be found in the colon.6 Common constitutional symptoms of colon GIST are similar to those of colon lipomas and include abdominal pain, changes in bowel habits, nausea, vomiting, and in some cases, weight loss.
CT imaging is often enough to differentiate a colon lipoma from a colon GIST. On CT, large GIST tumors tend to show irregular, lobulated margins, mucosal ulceration, central necrosis, cavitation, hemorrhage, and hypervascularity—vastly different from the CT findings of colon lipomas. If imaging is equivocal, an ultrasound-guided fine needle aspiration biopsy may be performed, differentiating GIST through the presence of ICC tumor cells as well as KIT and PDGFR-α proteins.
In our patient, colonoscopy showed a positive Cushion sign (tumor indented on depression with biopsy forceps), pathognomonic for a colon lipoma, and CT imaging showed a radiolucent, well-circumscribed lesion.9 This was more consistent with a colon lipoma than a GIST. Because the patient was symptomatic with a near obstructing lesion, the appropriate next step was removal of the lesion. Had this instead been a GIST tumor, a more extensive oncologic surgical resection would have been warranted, with adequate mesentery and lymph nodes collected.
This case is notable because colon lipomas exceeding 2 cm are rare and are usually an incidental finding on CT. However, larger lipomas can lead to symptoms, including obstruction if not removed in a timely manner.
1. Nallamothu G, Adler DG. Large colonic lipomas. Gastroenterol Hepatol (NY). 2011;7(7):490-492.
2. Crocetti D, Sapienza P, Sterpetti AV, et al. Surgery for symptomatic colon lipoma: a systematic review of the literature. Anticancer Res. 2014;34(11):6271-6276.
3. Italiano A, Ebran N, Attias R, et al. NFIB rearrangement in superficial, retroperitoneal, and colonic lipomas with aberrations involving chromosome band 9p22. Genes Chromosomes Cancer. 2008;47(11):971-977. doi:10.1002/gcc.20602
4. Agrawal A, Singh KJ. Symptomatic intestinal lipomas: our experience. Med J Armed Forces India. 2011;67(4):374-376. doi:10.1016/S0377-1237(11)60090-7
5. Kim GW, Kwon CI, Song SH, et al. Endoscopic resection of giant colonic lipoma: case series with partial resection. Clin Endosc. 2013;46(5):586-590. doi:10.5946/ce.2013.46.5.586
6. Reddy RM, Fleshman JW. Colorectal gastrointestinal stromal tumors: a brief review. Clin Colon Rectal Surg. 2006;19(2):69-77. doi:10.1055/s-2006-942347
7. Shinomura Y, Kinoshita K, Tsutsui S, Hirota S. Pathophysiology, diagnosis, and treatment of gastrointestinal stromal tumors. J Gastroenterol. 2005;40(8):775-780. doi:10.1007/s00535-005-1674-0
8. Patel N, Benipal B. Incidence of gastrointestinal stromal tumors in the United States from 2001-2015: a United States cancer statistics analysis of 50 states. Cureus. 2019;11(2):e4120. Published 2019 Feb 22. doi:10.7759/cureus.4120
9. Kyawzaw K, Emmanuel O, Sandar L,2 Febin J,Naing LA, Madhavi R. Pillow sign in colonoscopy. MOJ Clin Med Case Rep. 2018;8(2):57-58. doi:10.15406/mojcr.2018.08.00240
A 62-year-old male veteran with a history of pulmonary embolism (PE) and prostate cancer status after brachytherapy presented to the emergency department with new onset shortness of breath and left-sided chest pain after prolonged car travel. He underwent a chest computed tomography (CT) angiogram that showed no PE recurrence; however, the scan revealed an incidental transverse colon mass that appeared well circumscribed, homogeneous, and radiolucent with no enhancement, septations, or hypervascularity but no evidence of colonic distension or obstruction (Figure 1).
- What is your diagnosis?
- How would you treat this patient?
The patient reported having chronic constipation and a dull, left-sided abdominal discomfort for the past year. He noted that his abdominal pain worsened after eating and mildly improved after taking castor oil. He had no surgical history and no family history of cancer. The patient reported no fever, fatigue, weight loss, chills, nausea, vomiting, diarrhea, hematochezia, dysuria, hematuria, or melena. Vital signs, physical examination, and initial routine laboratory work were all within appropriate ranges, and a fecal occult blood test was negative.
A colonoscopy was performed, revealing a near-obstructing submucosal mass in the transverse colon near the splenic flexure with a smooth surface and a positive Cushion (Pillow) sign (Figure 2). The patient underwent surgical exploration that resulted in finding a soft, 11-cm lesion arising from the mesenteric side of the transverse colon (Figure 3). Hematoxylin and eosin (H&E) stains were used on a sample from the mass (Figure 4).
The tumor was enucleated via a colotomy over the mass, and the colotomy repaired primarily. Gross examination revealed homogenous yellow fatty tissue, and the H&E stains showed mature, well-differentiated adipocytes with uniform nuclei surrounded by a fibrous capsule. Based on this pathologic examination, this patient was diagnosed with a lipoma of the transverse colon. The resected tissue showed negative margins, indicating full removal of the lipoma.
The patient stabilized well after surgery and remained under inpatient care for observation; due to lack of appetite following the surgery, the patient did not start eating solids again until 2 days after the lipoma removal. By postoperative day 4, the patient had return of bowel function and was tolerating a regular diet with no recurrence of his prandial pain, shortness of breath, or left-sided chest pain. While the precise cause of the patient’s initial presentation of shortness of breath and left-sided chest pain was not ascertained, it is likely that the lipoma, near completely obstructed his bowel, caused abdominal contents and distended intestines to push against his diaphragm, leading to pain and dyspnea. This was likely exacerbated by sensitization to these symptoms from his prior PE. He was discharged home on postoperative day 4 with outpatient follow-up with general surgery.
Discussion
Lipomas are common benign tumors arising from aberrantly multiplying adipocytes. Although lipomas are most commonly found subcutaneously, the lesions can occur anywhere along the gastrointestinal (GI) tract, most often in the colon.1 The incidence rate of colon lipomas ranges from 0.2 to 4.4% among patients in their fifth to sixth decades of life, more commonly found in females.2 These lesions are the most common submucosal mesenchymal lesions of the colon, with a predilection for the right ascending colon.1 The etiology of colon lipomas is largely unknown; one known cause is trauma, thought to induce cytokine release or HMGA2-LPP fusion gene arrangements leading to adipocyte proliferation.3
Most colon lipomas are asymptomatic and discovered incidentally; symptoms typically arise when the lesions are > 2 cm in diameter and include abdominal pain, changes in bowel habits, rectal bleeding, and in extreme cases, obstruction and perforation.4 On CT imaging, colon lipomas will appear radiolucent, homogenous, and well circumscribed. The lesions usually do not warrant intervention unless they are symptomatic. If symptomatic, resection of the lesion is the first-line treatment and usually results in complete resolution of symptoms with no recurrence.2
While either a surgical or endoscopic approach may be used for resection, an increased risk of perforation of the colon with larger lipomas has been shown with endoscopic excision.5 With surgical resection, an open or minimally invasive approach may be offered, based on surgeon comfort with minimally invasive colon procedures. Minimally invasive colonic surgeries may be associated with a shorter length of stay, decreased postoperative pain, and faster return of bowel function. In this case, the surgeon chose an open approach due to the large size of the mass (11 cm) as well as location of the mass in the transverse colon, which made it easy to access directly through a small laparotomy incision made in the superior midline over the transverse colon.
When a colonic mesenchymal mass is seen on colonoscopy, it is important to consider other, nonbenign lesions that present this way. The most common malignant mesenchymal tumor of the GI tract is a gastrointestinal stromal tumor (GIST), a soft-tissue sarcoma that occurs predominantly in the stomach and small intestine.6 These tumors arise from the interstitial cells of Cajal (ICC) and are associated with mutations of KIT and PDGFR-α genes.7 The incidence in the United States is approximately 0.70 per 100,000 people per year, predominantly found in adults in their fifth or sixth decade of life.8 While this tumor typically occurs in the upper GI tract, very rarely, GISTs can be found in the colon.6 Common constitutional symptoms of colon GIST are similar to those of colon lipomas and include abdominal pain, changes in bowel habits, nausea, vomiting, and in some cases, weight loss.
CT imaging is often enough to differentiate a colon lipoma from a colon GIST. On CT, large GIST tumors tend to show irregular, lobulated margins, mucosal ulceration, central necrosis, cavitation, hemorrhage, and hypervascularity—vastly different from the CT findings of colon lipomas. If imaging is equivocal, an ultrasound-guided fine needle aspiration biopsy may be performed, differentiating GIST through the presence of ICC tumor cells as well as KIT and PDGFR-α proteins.
In our patient, colonoscopy showed a positive Cushion sign (tumor indented on depression with biopsy forceps), pathognomonic for a colon lipoma, and CT imaging showed a radiolucent, well-circumscribed lesion.9 This was more consistent with a colon lipoma than a GIST. Because the patient was symptomatic with a near obstructing lesion, the appropriate next step was removal of the lesion. Had this instead been a GIST tumor, a more extensive oncologic surgical resection would have been warranted, with adequate mesentery and lymph nodes collected.
This case is notable because colon lipomas exceeding 2 cm are rare and are usually an incidental finding on CT. However, larger lipomas can lead to symptoms, including obstruction if not removed in a timely manner.
A 62-year-old male veteran with a history of pulmonary embolism (PE) and prostate cancer status after brachytherapy presented to the emergency department with new onset shortness of breath and left-sided chest pain after prolonged car travel. He underwent a chest computed tomography (CT) angiogram that showed no PE recurrence; however, the scan revealed an incidental transverse colon mass that appeared well circumscribed, homogeneous, and radiolucent with no enhancement, septations, or hypervascularity but no evidence of colonic distension or obstruction (Figure 1).
- What is your diagnosis?
- How would you treat this patient?
The patient reported having chronic constipation and a dull, left-sided abdominal discomfort for the past year. He noted that his abdominal pain worsened after eating and mildly improved after taking castor oil. He had no surgical history and no family history of cancer. The patient reported no fever, fatigue, weight loss, chills, nausea, vomiting, diarrhea, hematochezia, dysuria, hematuria, or melena. Vital signs, physical examination, and initial routine laboratory work were all within appropriate ranges, and a fecal occult blood test was negative.
A colonoscopy was performed, revealing a near-obstructing submucosal mass in the transverse colon near the splenic flexure with a smooth surface and a positive Cushion (Pillow) sign (Figure 2). The patient underwent surgical exploration that resulted in finding a soft, 11-cm lesion arising from the mesenteric side of the transverse colon (Figure 3). Hematoxylin and eosin (H&E) stains were used on a sample from the mass (Figure 4).
The tumor was enucleated via a colotomy over the mass, and the colotomy repaired primarily. Gross examination revealed homogenous yellow fatty tissue, and the H&E stains showed mature, well-differentiated adipocytes with uniform nuclei surrounded by a fibrous capsule. Based on this pathologic examination, this patient was diagnosed with a lipoma of the transverse colon. The resected tissue showed negative margins, indicating full removal of the lipoma.
The patient stabilized well after surgery and remained under inpatient care for observation; due to lack of appetite following the surgery, the patient did not start eating solids again until 2 days after the lipoma removal. By postoperative day 4, the patient had return of bowel function and was tolerating a regular diet with no recurrence of his prandial pain, shortness of breath, or left-sided chest pain. While the precise cause of the patient’s initial presentation of shortness of breath and left-sided chest pain was not ascertained, it is likely that the lipoma, near completely obstructed his bowel, caused abdominal contents and distended intestines to push against his diaphragm, leading to pain and dyspnea. This was likely exacerbated by sensitization to these symptoms from his prior PE. He was discharged home on postoperative day 4 with outpatient follow-up with general surgery.
Discussion
Lipomas are common benign tumors arising from aberrantly multiplying adipocytes. Although lipomas are most commonly found subcutaneously, the lesions can occur anywhere along the gastrointestinal (GI) tract, most often in the colon.1 The incidence rate of colon lipomas ranges from 0.2 to 4.4% among patients in their fifth to sixth decades of life, more commonly found in females.2 These lesions are the most common submucosal mesenchymal lesions of the colon, with a predilection for the right ascending colon.1 The etiology of colon lipomas is largely unknown; one known cause is trauma, thought to induce cytokine release or HMGA2-LPP fusion gene arrangements leading to adipocyte proliferation.3
Most colon lipomas are asymptomatic and discovered incidentally; symptoms typically arise when the lesions are > 2 cm in diameter and include abdominal pain, changes in bowel habits, rectal bleeding, and in extreme cases, obstruction and perforation.4 On CT imaging, colon lipomas will appear radiolucent, homogenous, and well circumscribed. The lesions usually do not warrant intervention unless they are symptomatic. If symptomatic, resection of the lesion is the first-line treatment and usually results in complete resolution of symptoms with no recurrence.2
While either a surgical or endoscopic approach may be used for resection, an increased risk of perforation of the colon with larger lipomas has been shown with endoscopic excision.5 With surgical resection, an open or minimally invasive approach may be offered, based on surgeon comfort with minimally invasive colon procedures. Minimally invasive colonic surgeries may be associated with a shorter length of stay, decreased postoperative pain, and faster return of bowel function. In this case, the surgeon chose an open approach due to the large size of the mass (11 cm) as well as location of the mass in the transverse colon, which made it easy to access directly through a small laparotomy incision made in the superior midline over the transverse colon.
When a colonic mesenchymal mass is seen on colonoscopy, it is important to consider other, nonbenign lesions that present this way. The most common malignant mesenchymal tumor of the GI tract is a gastrointestinal stromal tumor (GIST), a soft-tissue sarcoma that occurs predominantly in the stomach and small intestine.6 These tumors arise from the interstitial cells of Cajal (ICC) and are associated with mutations of KIT and PDGFR-α genes.7 The incidence in the United States is approximately 0.70 per 100,000 people per year, predominantly found in adults in their fifth or sixth decade of life.8 While this tumor typically occurs in the upper GI tract, very rarely, GISTs can be found in the colon.6 Common constitutional symptoms of colon GIST are similar to those of colon lipomas and include abdominal pain, changes in bowel habits, nausea, vomiting, and in some cases, weight loss.
CT imaging is often enough to differentiate a colon lipoma from a colon GIST. On CT, large GIST tumors tend to show irregular, lobulated margins, mucosal ulceration, central necrosis, cavitation, hemorrhage, and hypervascularity—vastly different from the CT findings of colon lipomas. If imaging is equivocal, an ultrasound-guided fine needle aspiration biopsy may be performed, differentiating GIST through the presence of ICC tumor cells as well as KIT and PDGFR-α proteins.
In our patient, colonoscopy showed a positive Cushion sign (tumor indented on depression with biopsy forceps), pathognomonic for a colon lipoma, and CT imaging showed a radiolucent, well-circumscribed lesion.9 This was more consistent with a colon lipoma than a GIST. Because the patient was symptomatic with a near obstructing lesion, the appropriate next step was removal of the lesion. Had this instead been a GIST tumor, a more extensive oncologic surgical resection would have been warranted, with adequate mesentery and lymph nodes collected.
This case is notable because colon lipomas exceeding 2 cm are rare and are usually an incidental finding on CT. However, larger lipomas can lead to symptoms, including obstruction if not removed in a timely manner.
1. Nallamothu G, Adler DG. Large colonic lipomas. Gastroenterol Hepatol (NY). 2011;7(7):490-492.
2. Crocetti D, Sapienza P, Sterpetti AV, et al. Surgery for symptomatic colon lipoma: a systematic review of the literature. Anticancer Res. 2014;34(11):6271-6276.
3. Italiano A, Ebran N, Attias R, et al. NFIB rearrangement in superficial, retroperitoneal, and colonic lipomas with aberrations involving chromosome band 9p22. Genes Chromosomes Cancer. 2008;47(11):971-977. doi:10.1002/gcc.20602
4. Agrawal A, Singh KJ. Symptomatic intestinal lipomas: our experience. Med J Armed Forces India. 2011;67(4):374-376. doi:10.1016/S0377-1237(11)60090-7
5. Kim GW, Kwon CI, Song SH, et al. Endoscopic resection of giant colonic lipoma: case series with partial resection. Clin Endosc. 2013;46(5):586-590. doi:10.5946/ce.2013.46.5.586
6. Reddy RM, Fleshman JW. Colorectal gastrointestinal stromal tumors: a brief review. Clin Colon Rectal Surg. 2006;19(2):69-77. doi:10.1055/s-2006-942347
7. Shinomura Y, Kinoshita K, Tsutsui S, Hirota S. Pathophysiology, diagnosis, and treatment of gastrointestinal stromal tumors. J Gastroenterol. 2005;40(8):775-780. doi:10.1007/s00535-005-1674-0
8. Patel N, Benipal B. Incidence of gastrointestinal stromal tumors in the United States from 2001-2015: a United States cancer statistics analysis of 50 states. Cureus. 2019;11(2):e4120. Published 2019 Feb 22. doi:10.7759/cureus.4120
9. Kyawzaw K, Emmanuel O, Sandar L,2 Febin J,Naing LA, Madhavi R. Pillow sign in colonoscopy. MOJ Clin Med Case Rep. 2018;8(2):57-58. doi:10.15406/mojcr.2018.08.00240
1. Nallamothu G, Adler DG. Large colonic lipomas. Gastroenterol Hepatol (NY). 2011;7(7):490-492.
2. Crocetti D, Sapienza P, Sterpetti AV, et al. Surgery for symptomatic colon lipoma: a systematic review of the literature. Anticancer Res. 2014;34(11):6271-6276.
3. Italiano A, Ebran N, Attias R, et al. NFIB rearrangement in superficial, retroperitoneal, and colonic lipomas with aberrations involving chromosome band 9p22. Genes Chromosomes Cancer. 2008;47(11):971-977. doi:10.1002/gcc.20602
4. Agrawal A, Singh KJ. Symptomatic intestinal lipomas: our experience. Med J Armed Forces India. 2011;67(4):374-376. doi:10.1016/S0377-1237(11)60090-7
5. Kim GW, Kwon CI, Song SH, et al. Endoscopic resection of giant colonic lipoma: case series with partial resection. Clin Endosc. 2013;46(5):586-590. doi:10.5946/ce.2013.46.5.586
6. Reddy RM, Fleshman JW. Colorectal gastrointestinal stromal tumors: a brief review. Clin Colon Rectal Surg. 2006;19(2):69-77. doi:10.1055/s-2006-942347
7. Shinomura Y, Kinoshita K, Tsutsui S, Hirota S. Pathophysiology, diagnosis, and treatment of gastrointestinal stromal tumors. J Gastroenterol. 2005;40(8):775-780. doi:10.1007/s00535-005-1674-0
8. Patel N, Benipal B. Incidence of gastrointestinal stromal tumors in the United States from 2001-2015: a United States cancer statistics analysis of 50 states. Cureus. 2019;11(2):e4120. Published 2019 Feb 22. doi:10.7759/cureus.4120
9. Kyawzaw K, Emmanuel O, Sandar L,2 Febin J,Naing LA, Madhavi R. Pillow sign in colonoscopy. MOJ Clin Med Case Rep. 2018;8(2):57-58. doi:10.15406/mojcr.2018.08.00240
Patient Education After Inadequate Bowel Preparation: Improving Care and Outcomes
Colorectal cancer is the second most common cause of death in the United States. 1 A colonoscopy is the current gold standard for prevention and early detection of colorectal cancers. During a colonoscopy procedure, polyps and lesions are biopsied and removed. The most effective method of colon cleansing for the procedure is achieved by using one of several commercially available colon lavage preparations. Before the colonoscopy, patients are prescribed and instructed to take one of these bowel preparations.
Background
Adequate bowel preparation is defined as sufficient for identification of polyps > 5 mm.2 The impact of inadequate bowel preparation extends beyond the need for additional or repeat procedure(s) and includes potentially missed polyps and cancers. Inferior bowel preparation quality is associated with a significant decrease in the detection of flat or sessile serrated polyps.3 Missed polyps increase the risk of interval colorectal cancers. A high-quality bowel preparation together with the individual skill and experience of the endoscopist are crucial for adequate polyp detection. In addition, other risks of inadequate bowel preparation and repeated colonoscopies reduce adenoma detection rates, undetected carcinomas, and increase the risk of complications, possibly resulting in lawsuits.3
A major difficulty facing the Veterans Health Administration (VHA) medical center gastroenterologists is what to do when a patient is not properly prepared after standard prescreening education and the bowel preparation regimen. Traditionally, the patient is given additional medication and asked to return the next day for a repeat colonoscopy. Alternatively, the patient is given a 2-day bowel preparation to be used prior to a new appointment.
The choice of bowel preparation has been standardized within the US Department of Veterans Affairs (VA) Connecticut Healthcare System in West Haven (
For patients who fail the standard 1-day preparation, the same trained RNs inquire about any difficulties in consuming the preparation and provide the standard 2-day bowel preparation instructions. Multiple factors impact the adherence with preparation directions. Several patient-specific factors, comorbidities, and medications can contribute to inadequate bowel preparation.4 These factors include failing to fast before the procedure; namely, consuming solid foods, not consuming the entire preparation, not taking the preparation as directed, and not consuming adequate amounts of clear liquids or calories. Other reasons for failing the preparation are nausea and vomiting, poor understanding of instructions (including illiteracy), chronic constipation, use of narcotics and psychotropic drugs, and lack of awareness of the consequences of inadequate bowel preparation.
A study by Hautefeuille and colleagues noted that approximately 20% of patients having colonoscopy failure were not adherent to bowel preparation instructions.5 Only 55% of patients were aware of these consequences; whereas 96% of physicians were convinced they had given appropriate and sufficient information.5 As noted earlier, approximately half of patients do not fully comprehend the need to follow all the instructions. Therefore, clear and concise cleansing instructions and patient adherence are key factors that contribute to efficiency and quality of colonoscopy. The preparation failure rate creates a large volume of repeat patients and contributes to reduced efficiency of outpatient endoscopic practice.
A meta-analysis conducted by Chang and colleagues demonstrated that a brief counseling session with patients before colonoscopy ensured better bowel preparation.6 The focus of this article is on using the Colonoscopy Patient Education Bowel Preparation Questionnaire to improve the outcomes of patient education (Table).
As this was part of ongoing care and medication education; the research did not require reviews by a research committee or need institutional review board approval.
Questionnaire
A gastroenterology (GI) advance practice registered nurse (APRN) developed a patient questionnaire after reviewing patient records from 2016 through 2018 and noting information gaps in patient re-education. The information was not clearly and completely documented relating to frequency of bowel movements, constipation, and daily hydration/fluid intake. Several questions were consistently asked of patients who had previously failed 2 bowel preparations to determine the issues preventing a successful bowel cleansing. Notes from the GI and nutrition clinics and the primary care provider (PCP) were reviewed for information on constipation, frequency and quality of bowel movements, average beverage consumption, and hydration status.
The GI APRN conducted the review and used notes from the past year as well as the notes for prior colon preparations documenting bowel preparations and their resulting quality. A review was conducted on each patient who failed the standard 2-day bowel preparation before the GI APRN bowel preparation education session. The review revealed that no single note provided all necessary information. All colonoscopy prescreening education notes contained information from the standard prescreening preparation education class presentation, and any individual patient issues related to preparation consumption. GI and PCP notes included constipation information; however, frequency of bowel movements was seldom mentioned; and no fluid consumption information was provided except for alcohol related to abuse/addiction issues. Of the patients that had been seen by the Nutrition Department staff, their notes included caloric intake, appropriate food/dietary choices, and soda consumption; alcohol use was documented but related only to caloric intake; again, no other fluid intake amounts were documented.
Design
The questionnaire consists of 5 closed-ended, patient-centered questions aimed at accomplishing patient education in a time-efficient manner. It also is a tool to achieve consistency among staff in determining barriers and issues, improve documentation, and then assist the patient in achieving a good-to-excellent quality bowel preparation. The questions elicit information that allow an RN or PCP determine the factors that contributed to bowel preparation failure and allow for a tailored patient-education session. With a clear picture of the patient’s issues and obstacles, the patient-centered prescreening preparation education could focus on solutions to specific barriers, increase patient comprehension and adherence to the instructions, and identify complicating behavioral factors of the prior bowel preparation. For example, question 1 was designed to discover whether the patient failed to consume the preparation and why, such as volume, timing, or taste; question 4 was designed to assist in figuring out whether constipation for any reason may be present, whether currently diagnosed or not; and question 5 determined the risk of dehydration with or without constipation as a key cleansing issue.
The answers to these few questions determined whether the inadequate bowel preparation quality was due to issues of poor understanding, poor following of the directions, or to other complicating factors.
The prescreening bowel preparation education classes are delivered in groups classes, telehealth group classes, and by phone.
Discussion
Following implementation of the questionnaire from 2018 to 2019, a clinical chart review was conducted in 2019 of the first 100 patients who failed the standardized 2-day preparation from 2018 to 2019. These patients were selected by the GI attending physicians based on their multiple prior research studies and the total number of veterans served within VACHS to reflect an adequate test of change. Twenty patients canceled their appointments or refused to obtain an additional colonoscopy. Of the remaining 80 patients, 68 (85%) improved on the bowel preparation screening to an adequate rating.
Within the VACHS, the result of inadequate colon preparation leads to either an aborted colonoscopy or a longer examination duration due to time spent washing the colon mucosa and then suctioning the liquified stool. Using newchoicehealth.com 2021 national data, the colonoscopy average price range was $1800 to $12,500; the national average amount paid is $2750.7 The average screening or diagnostic colonoscopy cost was $4469.8
Using the Colonoscopy Patient-Education Bowel Prep Questionnaire resulted in increased patient satisfaction, better use of current patient appointment slots, increased unique encounters, and direct and indirect fiscal savings. Patient satisfaction resulted from no additional repeat colonoscopies per patient’s statements. The other findings resulted from the reduction in repeat appointments: The appointment slots that would have been taken by repeat colonoscopies were available for new patients, resulting in an increase in unique encounters.
Fiscal savings resulted from avoiding the need for additional bowel preparations for those patients or using the GI staff time (nurses and clerks) to reschedule and educate patients. Prior to the use of the questionnaire, patients who failed preparations would be re-educated, given a new preparation prescription or mailed a new preparation, scheduled, and then mailed the appropriate paperwork, thus, increasing the workload for nurses and clerks.
Conclusions
Use of the questionnaire resulted in increased high-quality bowel preparation, an increase in the number of unique patients served, and improved efficiency. In addition, recovered appointment slots and modest reductions in additional purchases of preparation kits resulted in a potential cost savings for VACHS. Proper cleansing instructions as well as identifying and overcoming barriers to achieving adequate preparation for colonoscopy resulted in improved patient satisfaction, quality care, and cost savings.
Regardless of the type of colon preparation, addressing patient barriers to bowel preparation is translatable to other endoscopy facilities and practices that provide patient education within the VA.
1. American Cancer Society. Key statistics for colorectal cancer. Revised January 12, 2021. Accessed May 19, 2021. https://www.cancer.org/cancer/colon-rectal-cancer/about/key-statistics.html. 2020.
2. Clark BT, Protiva P, Nagar A, et al. Quantification of adequate bowel preparation for screening or surveillance colonoscopy in men. Gastroenterology. 2016;150(2):396-405; quiz e14-e15. doi:10.1053/j.gastro.2015.09.041
3. Clark BT, Laine L. High-quality bowel preparation is required for detection of sessile serrated polyps. Clin Gastroenterol Hepatol. 2016;14(8):1155-1162. doi:10.1016/j.cgh.2016.03.044
4. D’Souza SM, Parekh PJ, Johnson DA. The dirty side of colonoscopy: predictors of poor bowel preparation and novel approaches to overcome the shortcomings. Br J Gastroenterology. 2019:1:1.
5. Hautefeuille G, Lapuelle J, Chaussade S, et al. Factors related to bowel cleansing failure before colonoscopy: results of the PACOME study. J United European Gastroenterol J. 2014; 2(1):22-29. doi:10.1177/2050640613518200
6. Chang CW, Shih SC, Wang HY, et al. Meta-analysis: the effect of patient education on bowel preparation for colonoscopy. Endosc Int Open. 2015;3(6):E646-E652. doi:10.1055/s-0034-1392365
7. New Choice Health. How much does a colonoscopy cost? Accessed May 19, 2021. https://www.newchoicehealth.com/colonoscopy/cost
8. MDsave.com. Colonoscopy. Accessed May 19, 2021. https://www.mdsave.com/f/procedure/colonoscopy/06516?q=colonoscopy&type=procedure
Colorectal cancer is the second most common cause of death in the United States. 1 A colonoscopy is the current gold standard for prevention and early detection of colorectal cancers. During a colonoscopy procedure, polyps and lesions are biopsied and removed. The most effective method of colon cleansing for the procedure is achieved by using one of several commercially available colon lavage preparations. Before the colonoscopy, patients are prescribed and instructed to take one of these bowel preparations.
Background
Adequate bowel preparation is defined as sufficient for identification of polyps > 5 mm.2 The impact of inadequate bowel preparation extends beyond the need for additional or repeat procedure(s) and includes potentially missed polyps and cancers. Inferior bowel preparation quality is associated with a significant decrease in the detection of flat or sessile serrated polyps.3 Missed polyps increase the risk of interval colorectal cancers. A high-quality bowel preparation together with the individual skill and experience of the endoscopist are crucial for adequate polyp detection. In addition, other risks of inadequate bowel preparation and repeated colonoscopies reduce adenoma detection rates, undetected carcinomas, and increase the risk of complications, possibly resulting in lawsuits.3
A major difficulty facing the Veterans Health Administration (VHA) medical center gastroenterologists is what to do when a patient is not properly prepared after standard prescreening education and the bowel preparation regimen. Traditionally, the patient is given additional medication and asked to return the next day for a repeat colonoscopy. Alternatively, the patient is given a 2-day bowel preparation to be used prior to a new appointment.
The choice of bowel preparation has been standardized within the US Department of Veterans Affairs (VA) Connecticut Healthcare System in West Haven (
For patients who fail the standard 1-day preparation, the same trained RNs inquire about any difficulties in consuming the preparation and provide the standard 2-day bowel preparation instructions. Multiple factors impact the adherence with preparation directions. Several patient-specific factors, comorbidities, and medications can contribute to inadequate bowel preparation.4 These factors include failing to fast before the procedure; namely, consuming solid foods, not consuming the entire preparation, not taking the preparation as directed, and not consuming adequate amounts of clear liquids or calories. Other reasons for failing the preparation are nausea and vomiting, poor understanding of instructions (including illiteracy), chronic constipation, use of narcotics and psychotropic drugs, and lack of awareness of the consequences of inadequate bowel preparation.
A study by Hautefeuille and colleagues noted that approximately 20% of patients having colonoscopy failure were not adherent to bowel preparation instructions.5 Only 55% of patients were aware of these consequences; whereas 96% of physicians were convinced they had given appropriate and sufficient information.5 As noted earlier, approximately half of patients do not fully comprehend the need to follow all the instructions. Therefore, clear and concise cleansing instructions and patient adherence are key factors that contribute to efficiency and quality of colonoscopy. The preparation failure rate creates a large volume of repeat patients and contributes to reduced efficiency of outpatient endoscopic practice.
A meta-analysis conducted by Chang and colleagues demonstrated that a brief counseling session with patients before colonoscopy ensured better bowel preparation.6 The focus of this article is on using the Colonoscopy Patient Education Bowel Preparation Questionnaire to improve the outcomes of patient education (Table).
As this was part of ongoing care and medication education; the research did not require reviews by a research committee or need institutional review board approval.
Questionnaire
A gastroenterology (GI) advance practice registered nurse (APRN) developed a patient questionnaire after reviewing patient records from 2016 through 2018 and noting information gaps in patient re-education. The information was not clearly and completely documented relating to frequency of bowel movements, constipation, and daily hydration/fluid intake. Several questions were consistently asked of patients who had previously failed 2 bowel preparations to determine the issues preventing a successful bowel cleansing. Notes from the GI and nutrition clinics and the primary care provider (PCP) were reviewed for information on constipation, frequency and quality of bowel movements, average beverage consumption, and hydration status.
The GI APRN conducted the review and used notes from the past year as well as the notes for prior colon preparations documenting bowel preparations and their resulting quality. A review was conducted on each patient who failed the standard 2-day bowel preparation before the GI APRN bowel preparation education session. The review revealed that no single note provided all necessary information. All colonoscopy prescreening education notes contained information from the standard prescreening preparation education class presentation, and any individual patient issues related to preparation consumption. GI and PCP notes included constipation information; however, frequency of bowel movements was seldom mentioned; and no fluid consumption information was provided except for alcohol related to abuse/addiction issues. Of the patients that had been seen by the Nutrition Department staff, their notes included caloric intake, appropriate food/dietary choices, and soda consumption; alcohol use was documented but related only to caloric intake; again, no other fluid intake amounts were documented.
Design
The questionnaire consists of 5 closed-ended, patient-centered questions aimed at accomplishing patient education in a time-efficient manner. It also is a tool to achieve consistency among staff in determining barriers and issues, improve documentation, and then assist the patient in achieving a good-to-excellent quality bowel preparation. The questions elicit information that allow an RN or PCP determine the factors that contributed to bowel preparation failure and allow for a tailored patient-education session. With a clear picture of the patient’s issues and obstacles, the patient-centered prescreening preparation education could focus on solutions to specific barriers, increase patient comprehension and adherence to the instructions, and identify complicating behavioral factors of the prior bowel preparation. For example, question 1 was designed to discover whether the patient failed to consume the preparation and why, such as volume, timing, or taste; question 4 was designed to assist in figuring out whether constipation for any reason may be present, whether currently diagnosed or not; and question 5 determined the risk of dehydration with or without constipation as a key cleansing issue.
The answers to these few questions determined whether the inadequate bowel preparation quality was due to issues of poor understanding, poor following of the directions, or to other complicating factors.
The prescreening bowel preparation education classes are delivered in groups classes, telehealth group classes, and by phone.
Discussion
Following implementation of the questionnaire from 2018 to 2019, a clinical chart review was conducted in 2019 of the first 100 patients who failed the standardized 2-day preparation from 2018 to 2019. These patients were selected by the GI attending physicians based on their multiple prior research studies and the total number of veterans served within VACHS to reflect an adequate test of change. Twenty patients canceled their appointments or refused to obtain an additional colonoscopy. Of the remaining 80 patients, 68 (85%) improved on the bowel preparation screening to an adequate rating.
Within the VACHS, the result of inadequate colon preparation leads to either an aborted colonoscopy or a longer examination duration due to time spent washing the colon mucosa and then suctioning the liquified stool. Using newchoicehealth.com 2021 national data, the colonoscopy average price range was $1800 to $12,500; the national average amount paid is $2750.7 The average screening or diagnostic colonoscopy cost was $4469.8
Using the Colonoscopy Patient-Education Bowel Prep Questionnaire resulted in increased patient satisfaction, better use of current patient appointment slots, increased unique encounters, and direct and indirect fiscal savings. Patient satisfaction resulted from no additional repeat colonoscopies per patient’s statements. The other findings resulted from the reduction in repeat appointments: The appointment slots that would have been taken by repeat colonoscopies were available for new patients, resulting in an increase in unique encounters.
Fiscal savings resulted from avoiding the need for additional bowel preparations for those patients or using the GI staff time (nurses and clerks) to reschedule and educate patients. Prior to the use of the questionnaire, patients who failed preparations would be re-educated, given a new preparation prescription or mailed a new preparation, scheduled, and then mailed the appropriate paperwork, thus, increasing the workload for nurses and clerks.
Conclusions
Use of the questionnaire resulted in increased high-quality bowel preparation, an increase in the number of unique patients served, and improved efficiency. In addition, recovered appointment slots and modest reductions in additional purchases of preparation kits resulted in a potential cost savings for VACHS. Proper cleansing instructions as well as identifying and overcoming barriers to achieving adequate preparation for colonoscopy resulted in improved patient satisfaction, quality care, and cost savings.
Regardless of the type of colon preparation, addressing patient barriers to bowel preparation is translatable to other endoscopy facilities and practices that provide patient education within the VA.
Colorectal cancer is the second most common cause of death in the United States. 1 A colonoscopy is the current gold standard for prevention and early detection of colorectal cancers. During a colonoscopy procedure, polyps and lesions are biopsied and removed. The most effective method of colon cleansing for the procedure is achieved by using one of several commercially available colon lavage preparations. Before the colonoscopy, patients are prescribed and instructed to take one of these bowel preparations.
Background
Adequate bowel preparation is defined as sufficient for identification of polyps > 5 mm.2 The impact of inadequate bowel preparation extends beyond the need for additional or repeat procedure(s) and includes potentially missed polyps and cancers. Inferior bowel preparation quality is associated with a significant decrease in the detection of flat or sessile serrated polyps.3 Missed polyps increase the risk of interval colorectal cancers. A high-quality bowel preparation together with the individual skill and experience of the endoscopist are crucial for adequate polyp detection. In addition, other risks of inadequate bowel preparation and repeated colonoscopies reduce adenoma detection rates, undetected carcinomas, and increase the risk of complications, possibly resulting in lawsuits.3
A major difficulty facing the Veterans Health Administration (VHA) medical center gastroenterologists is what to do when a patient is not properly prepared after standard prescreening education and the bowel preparation regimen. Traditionally, the patient is given additional medication and asked to return the next day for a repeat colonoscopy. Alternatively, the patient is given a 2-day bowel preparation to be used prior to a new appointment.
The choice of bowel preparation has been standardized within the US Department of Veterans Affairs (VA) Connecticut Healthcare System in West Haven (
For patients who fail the standard 1-day preparation, the same trained RNs inquire about any difficulties in consuming the preparation and provide the standard 2-day bowel preparation instructions. Multiple factors impact the adherence with preparation directions. Several patient-specific factors, comorbidities, and medications can contribute to inadequate bowel preparation.4 These factors include failing to fast before the procedure; namely, consuming solid foods, not consuming the entire preparation, not taking the preparation as directed, and not consuming adequate amounts of clear liquids or calories. Other reasons for failing the preparation are nausea and vomiting, poor understanding of instructions (including illiteracy), chronic constipation, use of narcotics and psychotropic drugs, and lack of awareness of the consequences of inadequate bowel preparation.
A study by Hautefeuille and colleagues noted that approximately 20% of patients having colonoscopy failure were not adherent to bowel preparation instructions.5 Only 55% of patients were aware of these consequences; whereas 96% of physicians were convinced they had given appropriate and sufficient information.5 As noted earlier, approximately half of patients do not fully comprehend the need to follow all the instructions. Therefore, clear and concise cleansing instructions and patient adherence are key factors that contribute to efficiency and quality of colonoscopy. The preparation failure rate creates a large volume of repeat patients and contributes to reduced efficiency of outpatient endoscopic practice.
A meta-analysis conducted by Chang and colleagues demonstrated that a brief counseling session with patients before colonoscopy ensured better bowel preparation.6 The focus of this article is on using the Colonoscopy Patient Education Bowel Preparation Questionnaire to improve the outcomes of patient education (Table).
As this was part of ongoing care and medication education; the research did not require reviews by a research committee or need institutional review board approval.
Questionnaire
A gastroenterology (GI) advance practice registered nurse (APRN) developed a patient questionnaire after reviewing patient records from 2016 through 2018 and noting information gaps in patient re-education. The information was not clearly and completely documented relating to frequency of bowel movements, constipation, and daily hydration/fluid intake. Several questions were consistently asked of patients who had previously failed 2 bowel preparations to determine the issues preventing a successful bowel cleansing. Notes from the GI and nutrition clinics and the primary care provider (PCP) were reviewed for information on constipation, frequency and quality of bowel movements, average beverage consumption, and hydration status.
The GI APRN conducted the review and used notes from the past year as well as the notes for prior colon preparations documenting bowel preparations and their resulting quality. A review was conducted on each patient who failed the standard 2-day bowel preparation before the GI APRN bowel preparation education session. The review revealed that no single note provided all necessary information. All colonoscopy prescreening education notes contained information from the standard prescreening preparation education class presentation, and any individual patient issues related to preparation consumption. GI and PCP notes included constipation information; however, frequency of bowel movements was seldom mentioned; and no fluid consumption information was provided except for alcohol related to abuse/addiction issues. Of the patients that had been seen by the Nutrition Department staff, their notes included caloric intake, appropriate food/dietary choices, and soda consumption; alcohol use was documented but related only to caloric intake; again, no other fluid intake amounts were documented.
Design
The questionnaire consists of 5 closed-ended, patient-centered questions aimed at accomplishing patient education in a time-efficient manner. It also is a tool to achieve consistency among staff in determining barriers and issues, improve documentation, and then assist the patient in achieving a good-to-excellent quality bowel preparation. The questions elicit information that allow an RN or PCP determine the factors that contributed to bowel preparation failure and allow for a tailored patient-education session. With a clear picture of the patient’s issues and obstacles, the patient-centered prescreening preparation education could focus on solutions to specific barriers, increase patient comprehension and adherence to the instructions, and identify complicating behavioral factors of the prior bowel preparation. For example, question 1 was designed to discover whether the patient failed to consume the preparation and why, such as volume, timing, or taste; question 4 was designed to assist in figuring out whether constipation for any reason may be present, whether currently diagnosed or not; and question 5 determined the risk of dehydration with or without constipation as a key cleansing issue.
The answers to these few questions determined whether the inadequate bowel preparation quality was due to issues of poor understanding, poor following of the directions, or to other complicating factors.
The prescreening bowel preparation education classes are delivered in groups classes, telehealth group classes, and by phone.
Discussion
Following implementation of the questionnaire from 2018 to 2019, a clinical chart review was conducted in 2019 of the first 100 patients who failed the standardized 2-day preparation from 2018 to 2019. These patients were selected by the GI attending physicians based on their multiple prior research studies and the total number of veterans served within VACHS to reflect an adequate test of change. Twenty patients canceled their appointments or refused to obtain an additional colonoscopy. Of the remaining 80 patients, 68 (85%) improved on the bowel preparation screening to an adequate rating.
Within the VACHS, the result of inadequate colon preparation leads to either an aborted colonoscopy or a longer examination duration due to time spent washing the colon mucosa and then suctioning the liquified stool. Using newchoicehealth.com 2021 national data, the colonoscopy average price range was $1800 to $12,500; the national average amount paid is $2750.7 The average screening or diagnostic colonoscopy cost was $4469.8
Using the Colonoscopy Patient-Education Bowel Prep Questionnaire resulted in increased patient satisfaction, better use of current patient appointment slots, increased unique encounters, and direct and indirect fiscal savings. Patient satisfaction resulted from no additional repeat colonoscopies per patient’s statements. The other findings resulted from the reduction in repeat appointments: The appointment slots that would have been taken by repeat colonoscopies were available for new patients, resulting in an increase in unique encounters.
Fiscal savings resulted from avoiding the need for additional bowel preparations for those patients or using the GI staff time (nurses and clerks) to reschedule and educate patients. Prior to the use of the questionnaire, patients who failed preparations would be re-educated, given a new preparation prescription or mailed a new preparation, scheduled, and then mailed the appropriate paperwork, thus, increasing the workload for nurses and clerks.
Conclusions
Use of the questionnaire resulted in increased high-quality bowel preparation, an increase in the number of unique patients served, and improved efficiency. In addition, recovered appointment slots and modest reductions in additional purchases of preparation kits resulted in a potential cost savings for VACHS. Proper cleansing instructions as well as identifying and overcoming barriers to achieving adequate preparation for colonoscopy resulted in improved patient satisfaction, quality care, and cost savings.
Regardless of the type of colon preparation, addressing patient barriers to bowel preparation is translatable to other endoscopy facilities and practices that provide patient education within the VA.
1. American Cancer Society. Key statistics for colorectal cancer. Revised January 12, 2021. Accessed May 19, 2021. https://www.cancer.org/cancer/colon-rectal-cancer/about/key-statistics.html. 2020.
2. Clark BT, Protiva P, Nagar A, et al. Quantification of adequate bowel preparation for screening or surveillance colonoscopy in men. Gastroenterology. 2016;150(2):396-405; quiz e14-e15. doi:10.1053/j.gastro.2015.09.041
3. Clark BT, Laine L. High-quality bowel preparation is required for detection of sessile serrated polyps. Clin Gastroenterol Hepatol. 2016;14(8):1155-1162. doi:10.1016/j.cgh.2016.03.044
4. D’Souza SM, Parekh PJ, Johnson DA. The dirty side of colonoscopy: predictors of poor bowel preparation and novel approaches to overcome the shortcomings. Br J Gastroenterology. 2019:1:1.
5. Hautefeuille G, Lapuelle J, Chaussade S, et al. Factors related to bowel cleansing failure before colonoscopy: results of the PACOME study. J United European Gastroenterol J. 2014; 2(1):22-29. doi:10.1177/2050640613518200
6. Chang CW, Shih SC, Wang HY, et al. Meta-analysis: the effect of patient education on bowel preparation for colonoscopy. Endosc Int Open. 2015;3(6):E646-E652. doi:10.1055/s-0034-1392365
7. New Choice Health. How much does a colonoscopy cost? Accessed May 19, 2021. https://www.newchoicehealth.com/colonoscopy/cost
8. MDsave.com. Colonoscopy. Accessed May 19, 2021. https://www.mdsave.com/f/procedure/colonoscopy/06516?q=colonoscopy&type=procedure
1. American Cancer Society. Key statistics for colorectal cancer. Revised January 12, 2021. Accessed May 19, 2021. https://www.cancer.org/cancer/colon-rectal-cancer/about/key-statistics.html. 2020.
2. Clark BT, Protiva P, Nagar A, et al. Quantification of adequate bowel preparation for screening or surveillance colonoscopy in men. Gastroenterology. 2016;150(2):396-405; quiz e14-e15. doi:10.1053/j.gastro.2015.09.041
3. Clark BT, Laine L. High-quality bowel preparation is required for detection of sessile serrated polyps. Clin Gastroenterol Hepatol. 2016;14(8):1155-1162. doi:10.1016/j.cgh.2016.03.044
4. D’Souza SM, Parekh PJ, Johnson DA. The dirty side of colonoscopy: predictors of poor bowel preparation and novel approaches to overcome the shortcomings. Br J Gastroenterology. 2019:1:1.
5. Hautefeuille G, Lapuelle J, Chaussade S, et al. Factors related to bowel cleansing failure before colonoscopy: results of the PACOME study. J United European Gastroenterol J. 2014; 2(1):22-29. doi:10.1177/2050640613518200
6. Chang CW, Shih SC, Wang HY, et al. Meta-analysis: the effect of patient education on bowel preparation for colonoscopy. Endosc Int Open. 2015;3(6):E646-E652. doi:10.1055/s-0034-1392365
7. New Choice Health. How much does a colonoscopy cost? Accessed May 19, 2021. https://www.newchoicehealth.com/colonoscopy/cost
8. MDsave.com. Colonoscopy. Accessed May 19, 2021. https://www.mdsave.com/f/procedure/colonoscopy/06516?q=colonoscopy&type=procedure
Chemotherapy/local excision avoids proctectomy in rectal cancer
Chemotherapy and local excision led to organ preservation in over half of early-stage rectal cancer patients in a small study, but follow-up was only a median of 15.4 months.
Even so, “we believe that subsequent trials ... are warranted,” said lead investigator Hagen F. Kennecke, MD, medical director of GI oncology at Providence Cancer Institute, Portland, Ore., who presented the findings at the American Society of Clinical Oncology annual meeting.
“The results are quite promising,” said study discussant Karyn Stitzenberg, MD, a surgical oncologist at the University of North Carolina, Chapel Hill.
“The reported organ preservation rates of 57% to 79% compare favorably with the rates previously demonstrated in studies of neoadjuvant chemoradiation followed by local excision,” but longer-term follow up is needed “to know the true organ preservation rate,” she said.
Organ preservation – sparing the rectum during treatment – is a hot topic in rectal cancer. Total mesorectal excision (TME) is still the go-to option, but it’s fraught with bad GI, urinary, sexual, and other complications for patients. “Consequently, the concept of organ preservation ... is very appealing,” Dr. Stitzenberg explained.
The chemoradiation/local excision approach she referenced is gaining traction as an alternative, but the radiation component is itself associated with substantial short- and long-term problems, including sphincter dysfunction and wound healing complications.
The goal of Dr. Kennecke’s study, dubbed NEO [Neoadjuvant, Excision, Observation], was to see if the radiation could be left out altogether.
Recruited at eight centers in Canada and one in the United States, the 58 subjects had clinical stage T1-T3 A/B node-negative tumors with no pathologic high-risk features.
They received neoadjuvant FOLFOX (six cycles in 32 patients, 91% completion rate) or CAPOX (four cycles in 26 patients, 89% completion); 56 of the 58 subjects then went on to transanal endoscopic tumor excision; one of the other two patients wasn’t eligible because of tumor progression and the other one declined.
The 33 patients who were stage T0/T1N0 after treatment were spared organ removal and underwent observation every 3-6 months. TME was recommended for the 23 others who were stage 2 or higher or had nodal metastases following chemotherapy and excision.
The numbers translated to a per-protocol organ preservation rate of 57% over a median follow-up of 15.4 months; when the 13 patients who declined TME were added, the rate climbed to 79%.
Although “organ preservation in rectal cancer is becoming an increasingly promising and realistic option for a subset of patients,” Dr. Stitzenberg said, there are more reasons to be cautious beyond the short follow-up.
“The standard of care treatment for these patients would have been proctectomy ... Most would not have [had] systemic chemotherapy. As a result, the added morbidity of FOLFOX or CAPOX needs to be considered.” The study reported that there were no unexpected toxicities, but “what were the expected toxicities? How many patients experienced grade 3 to 5 complications?” she wondered.
Also, how realistic is it to expect patients to report for surveillance every few months outside of a trial? And how can they best be watched to make sure recurrence is caught “while salvage TME is still feasible? There are many longer-term follow-up questions that remain to be answered,” Dr. Stitzenberg said.
Even with short follow-up, there were two locoregional recurrences across the cohort (3.5%), both treated by TME to R0/1 resection. There were no distant relapses.
Subjects were a median of 67 years old, and over two-thirds were men. The majority had stage 2 disease at baseline. Tumors were well to moderately differentiated nonmucinous rectal adenocarcinomas with a median height of 6 cm.
The work was funded by the Canadian Cancer Trials Group. Dr. Kennecke disclosed relationships with Advanced Accelerator Applications, Ipsen, and Taiho Pharmaceutical. Dr. Stitzenberg had no relevant disclosures.
Chemotherapy and local excision led to organ preservation in over half of early-stage rectal cancer patients in a small study, but follow-up was only a median of 15.4 months.
Even so, “we believe that subsequent trials ... are warranted,” said lead investigator Hagen F. Kennecke, MD, medical director of GI oncology at Providence Cancer Institute, Portland, Ore., who presented the findings at the American Society of Clinical Oncology annual meeting.
“The results are quite promising,” said study discussant Karyn Stitzenberg, MD, a surgical oncologist at the University of North Carolina, Chapel Hill.
“The reported organ preservation rates of 57% to 79% compare favorably with the rates previously demonstrated in studies of neoadjuvant chemoradiation followed by local excision,” but longer-term follow up is needed “to know the true organ preservation rate,” she said.
Organ preservation – sparing the rectum during treatment – is a hot topic in rectal cancer. Total mesorectal excision (TME) is still the go-to option, but it’s fraught with bad GI, urinary, sexual, and other complications for patients. “Consequently, the concept of organ preservation ... is very appealing,” Dr. Stitzenberg explained.
The chemoradiation/local excision approach she referenced is gaining traction as an alternative, but the radiation component is itself associated with substantial short- and long-term problems, including sphincter dysfunction and wound healing complications.
The goal of Dr. Kennecke’s study, dubbed NEO [Neoadjuvant, Excision, Observation], was to see if the radiation could be left out altogether.
Recruited at eight centers in Canada and one in the United States, the 58 subjects had clinical stage T1-T3 A/B node-negative tumors with no pathologic high-risk features.
They received neoadjuvant FOLFOX (six cycles in 32 patients, 91% completion rate) or CAPOX (four cycles in 26 patients, 89% completion); 56 of the 58 subjects then went on to transanal endoscopic tumor excision; one of the other two patients wasn’t eligible because of tumor progression and the other one declined.
The 33 patients who were stage T0/T1N0 after treatment were spared organ removal and underwent observation every 3-6 months. TME was recommended for the 23 others who were stage 2 or higher or had nodal metastases following chemotherapy and excision.
The numbers translated to a per-protocol organ preservation rate of 57% over a median follow-up of 15.4 months; when the 13 patients who declined TME were added, the rate climbed to 79%.
Although “organ preservation in rectal cancer is becoming an increasingly promising and realistic option for a subset of patients,” Dr. Stitzenberg said, there are more reasons to be cautious beyond the short follow-up.
“The standard of care treatment for these patients would have been proctectomy ... Most would not have [had] systemic chemotherapy. As a result, the added morbidity of FOLFOX or CAPOX needs to be considered.” The study reported that there were no unexpected toxicities, but “what were the expected toxicities? How many patients experienced grade 3 to 5 complications?” she wondered.
Also, how realistic is it to expect patients to report for surveillance every few months outside of a trial? And how can they best be watched to make sure recurrence is caught “while salvage TME is still feasible? There are many longer-term follow-up questions that remain to be answered,” Dr. Stitzenberg said.
Even with short follow-up, there were two locoregional recurrences across the cohort (3.5%), both treated by TME to R0/1 resection. There were no distant relapses.
Subjects were a median of 67 years old, and over two-thirds were men. The majority had stage 2 disease at baseline. Tumors were well to moderately differentiated nonmucinous rectal adenocarcinomas with a median height of 6 cm.
The work was funded by the Canadian Cancer Trials Group. Dr. Kennecke disclosed relationships with Advanced Accelerator Applications, Ipsen, and Taiho Pharmaceutical. Dr. Stitzenberg had no relevant disclosures.
Chemotherapy and local excision led to organ preservation in over half of early-stage rectal cancer patients in a small study, but follow-up was only a median of 15.4 months.
Even so, “we believe that subsequent trials ... are warranted,” said lead investigator Hagen F. Kennecke, MD, medical director of GI oncology at Providence Cancer Institute, Portland, Ore., who presented the findings at the American Society of Clinical Oncology annual meeting.
“The results are quite promising,” said study discussant Karyn Stitzenberg, MD, a surgical oncologist at the University of North Carolina, Chapel Hill.
“The reported organ preservation rates of 57% to 79% compare favorably with the rates previously demonstrated in studies of neoadjuvant chemoradiation followed by local excision,” but longer-term follow up is needed “to know the true organ preservation rate,” she said.
Organ preservation – sparing the rectum during treatment – is a hot topic in rectal cancer. Total mesorectal excision (TME) is still the go-to option, but it’s fraught with bad GI, urinary, sexual, and other complications for patients. “Consequently, the concept of organ preservation ... is very appealing,” Dr. Stitzenberg explained.
The chemoradiation/local excision approach she referenced is gaining traction as an alternative, but the radiation component is itself associated with substantial short- and long-term problems, including sphincter dysfunction and wound healing complications.
The goal of Dr. Kennecke’s study, dubbed NEO [Neoadjuvant, Excision, Observation], was to see if the radiation could be left out altogether.
Recruited at eight centers in Canada and one in the United States, the 58 subjects had clinical stage T1-T3 A/B node-negative tumors with no pathologic high-risk features.
They received neoadjuvant FOLFOX (six cycles in 32 patients, 91% completion rate) or CAPOX (four cycles in 26 patients, 89% completion); 56 of the 58 subjects then went on to transanal endoscopic tumor excision; one of the other two patients wasn’t eligible because of tumor progression and the other one declined.
The 33 patients who were stage T0/T1N0 after treatment were spared organ removal and underwent observation every 3-6 months. TME was recommended for the 23 others who were stage 2 or higher or had nodal metastases following chemotherapy and excision.
The numbers translated to a per-protocol organ preservation rate of 57% over a median follow-up of 15.4 months; when the 13 patients who declined TME were added, the rate climbed to 79%.
Although “organ preservation in rectal cancer is becoming an increasingly promising and realistic option for a subset of patients,” Dr. Stitzenberg said, there are more reasons to be cautious beyond the short follow-up.
“The standard of care treatment for these patients would have been proctectomy ... Most would not have [had] systemic chemotherapy. As a result, the added morbidity of FOLFOX or CAPOX needs to be considered.” The study reported that there were no unexpected toxicities, but “what were the expected toxicities? How many patients experienced grade 3 to 5 complications?” she wondered.
Also, how realistic is it to expect patients to report for surveillance every few months outside of a trial? And how can they best be watched to make sure recurrence is caught “while salvage TME is still feasible? There are many longer-term follow-up questions that remain to be answered,” Dr. Stitzenberg said.
Even with short follow-up, there were two locoregional recurrences across the cohort (3.5%), both treated by TME to R0/1 resection. There were no distant relapses.
Subjects were a median of 67 years old, and over two-thirds were men. The majority had stage 2 disease at baseline. Tumors were well to moderately differentiated nonmucinous rectal adenocarcinomas with a median height of 6 cm.
The work was funded by the Canadian Cancer Trials Group. Dr. Kennecke disclosed relationships with Advanced Accelerator Applications, Ipsen, and Taiho Pharmaceutical. Dr. Stitzenberg had no relevant disclosures.
FROM ASCO 2021
Upfront asymptomatic primary tumor resection: No benefit in advanced CRC
Upfront resection of an asymptomatic primary tumor does not improve survival over chemotherapy alone in stage IV colorectal cancer with unresectable synchronous metastases, according to a randomized trial in Japan with 165 patients.
Median overall survival was slightly more than 2 years with or without primary resection, plus upfront surgery delayed systemic treatment and had a 4% risk of fatal complications. The trial was terminated early because of futility.
“PTR [primary tumor resection] should no longer be considered a standard of care for patients with CRC with asymptomatic primary tumors and synchronous unresectable metastases,” said investigators led by Yukihide Kanemitsu, MD, a colorectal surgeon at National Cancer Center Hospital in Tokyo.
“This paper is important for establishing solid evidence-based decisions. Why perform an invasive procedure on a patient that introduces additional risks if it won’t change their disease course?” said colorectal surgeon Deborah Keller, MD, clinical assistant professor of surgery at the University of California at Davis Medical Center, when asked for comment.
She explained that, in general, when the primary tumor is not obstructing, the standard of care is upfront chemotherapy, as supported by National Comprehensive Cancer Network guidelines.
“However, there was a change in practice over the last few years” after several retrospective studies reported better overall survival with surgery before chemotherapy, but “the studies were not the highest quality of evidence,” she said.
To bring better data to bear, the Japanese team randomized 84 patients to chemotherapy alone and 81 to PTR followed by chemotherapy. Primary tumors were asymptomatic, and patients had no more than three unresectable metastases in the liver, lungs, distant lymph nodes, or peritoneum. Chemotherapy was either mFOLFOX6 plus bevacizumab or CapeOX plus bevacizumab at investigator discretion.
The trial was halted at interim analysis in September 2019. With a median follow-up of 22 months, median overall survival – the primary endpoint – was 25.9 months in the surgery-first arm and 26.7 months in the chemotherapy-alone group (P = .69). Subgroup analysis found no populations that benefited from PTR. Median progression free survival was 10.4 months with PTR first and 12.1 months with chemotherapy alone (P = .48)
Twenty-seven patients in the PTR arm had grade 3 or worse surgery-related adverse events, including anastomotic leakage in 3 and increased aspartate aminotransferase in 13. Three patients (4%) died within 30 days of surgery. Chemotherapy-related grade 3 or worse adverse events were more frequent and severe in the PTR arm (48% PTR versus 34% chemotherapy alone).
“For those who had been performing resections, this could push the paradigm back to chemotherapy alone,” Dr. Keller said.
Eleven patients (13%) in the chemotherapy-alone arm required surgery for intestinal obstruction or other primary tumor symptoms that developed after randomization, which means that 87% avoided surgery entirely, the investigators noted.
In the PTR group, surgery was performed within 21 days of enrollment, and chemotherapy started a median of 34 days after PTR. Chemotherapy was started within 14 days of enrollment in the chemotherapy-alone arm.
The groups were well balanced, including colon and rectosigmoid tumor locations in 93% of both arms and liver metastases in 73% of both. A bit over half the subjects were men and the median age was 65 years.
The team noted that early termination meant that the planned sample size was not achieved, which in turn limited the statistical power of the conclusions. “It is hoped that the comprehensive results of [ongoing trials] will clearly demonstrate the role of PTR for these patients,” they said.
The study was conducted by the Japan Clinical Oncology Group at 38 cancer centers in Japan. The work was funded by the Ministry of Health of Japan. The investigators had numerous industry ties, including Dr. Kanemitsu, who reported honoraria from Chugai Pharma, Ethicon, Covidien, and Intuitive Surgical, and being a Covidien adviser.
Upfront resection of an asymptomatic primary tumor does not improve survival over chemotherapy alone in stage IV colorectal cancer with unresectable synchronous metastases, according to a randomized trial in Japan with 165 patients.
Median overall survival was slightly more than 2 years with or without primary resection, plus upfront surgery delayed systemic treatment and had a 4% risk of fatal complications. The trial was terminated early because of futility.
“PTR [primary tumor resection] should no longer be considered a standard of care for patients with CRC with asymptomatic primary tumors and synchronous unresectable metastases,” said investigators led by Yukihide Kanemitsu, MD, a colorectal surgeon at National Cancer Center Hospital in Tokyo.
“This paper is important for establishing solid evidence-based decisions. Why perform an invasive procedure on a patient that introduces additional risks if it won’t change their disease course?” said colorectal surgeon Deborah Keller, MD, clinical assistant professor of surgery at the University of California at Davis Medical Center, when asked for comment.
She explained that, in general, when the primary tumor is not obstructing, the standard of care is upfront chemotherapy, as supported by National Comprehensive Cancer Network guidelines.
“However, there was a change in practice over the last few years” after several retrospective studies reported better overall survival with surgery before chemotherapy, but “the studies were not the highest quality of evidence,” she said.
To bring better data to bear, the Japanese team randomized 84 patients to chemotherapy alone and 81 to PTR followed by chemotherapy. Primary tumors were asymptomatic, and patients had no more than three unresectable metastases in the liver, lungs, distant lymph nodes, or peritoneum. Chemotherapy was either mFOLFOX6 plus bevacizumab or CapeOX plus bevacizumab at investigator discretion.
The trial was halted at interim analysis in September 2019. With a median follow-up of 22 months, median overall survival – the primary endpoint – was 25.9 months in the surgery-first arm and 26.7 months in the chemotherapy-alone group (P = .69). Subgroup analysis found no populations that benefited from PTR. Median progression free survival was 10.4 months with PTR first and 12.1 months with chemotherapy alone (P = .48)
Twenty-seven patients in the PTR arm had grade 3 or worse surgery-related adverse events, including anastomotic leakage in 3 and increased aspartate aminotransferase in 13. Three patients (4%) died within 30 days of surgery. Chemotherapy-related grade 3 or worse adverse events were more frequent and severe in the PTR arm (48% PTR versus 34% chemotherapy alone).
“For those who had been performing resections, this could push the paradigm back to chemotherapy alone,” Dr. Keller said.
Eleven patients (13%) in the chemotherapy-alone arm required surgery for intestinal obstruction or other primary tumor symptoms that developed after randomization, which means that 87% avoided surgery entirely, the investigators noted.
In the PTR group, surgery was performed within 21 days of enrollment, and chemotherapy started a median of 34 days after PTR. Chemotherapy was started within 14 days of enrollment in the chemotherapy-alone arm.
The groups were well balanced, including colon and rectosigmoid tumor locations in 93% of both arms and liver metastases in 73% of both. A bit over half the subjects were men and the median age was 65 years.
The team noted that early termination meant that the planned sample size was not achieved, which in turn limited the statistical power of the conclusions. “It is hoped that the comprehensive results of [ongoing trials] will clearly demonstrate the role of PTR for these patients,” they said.
The study was conducted by the Japan Clinical Oncology Group at 38 cancer centers in Japan. The work was funded by the Ministry of Health of Japan. The investigators had numerous industry ties, including Dr. Kanemitsu, who reported honoraria from Chugai Pharma, Ethicon, Covidien, and Intuitive Surgical, and being a Covidien adviser.
Upfront resection of an asymptomatic primary tumor does not improve survival over chemotherapy alone in stage IV colorectal cancer with unresectable synchronous metastases, according to a randomized trial in Japan with 165 patients.
Median overall survival was slightly more than 2 years with or without primary resection, plus upfront surgery delayed systemic treatment and had a 4% risk of fatal complications. The trial was terminated early because of futility.
“PTR [primary tumor resection] should no longer be considered a standard of care for patients with CRC with asymptomatic primary tumors and synchronous unresectable metastases,” said investigators led by Yukihide Kanemitsu, MD, a colorectal surgeon at National Cancer Center Hospital in Tokyo.
“This paper is important for establishing solid evidence-based decisions. Why perform an invasive procedure on a patient that introduces additional risks if it won’t change their disease course?” said colorectal surgeon Deborah Keller, MD, clinical assistant professor of surgery at the University of California at Davis Medical Center, when asked for comment.
She explained that, in general, when the primary tumor is not obstructing, the standard of care is upfront chemotherapy, as supported by National Comprehensive Cancer Network guidelines.
“However, there was a change in practice over the last few years” after several retrospective studies reported better overall survival with surgery before chemotherapy, but “the studies were not the highest quality of evidence,” she said.
To bring better data to bear, the Japanese team randomized 84 patients to chemotherapy alone and 81 to PTR followed by chemotherapy. Primary tumors were asymptomatic, and patients had no more than three unresectable metastases in the liver, lungs, distant lymph nodes, or peritoneum. Chemotherapy was either mFOLFOX6 plus bevacizumab or CapeOX plus bevacizumab at investigator discretion.
The trial was halted at interim analysis in September 2019. With a median follow-up of 22 months, median overall survival – the primary endpoint – was 25.9 months in the surgery-first arm and 26.7 months in the chemotherapy-alone group (P = .69). Subgroup analysis found no populations that benefited from PTR. Median progression free survival was 10.4 months with PTR first and 12.1 months with chemotherapy alone (P = .48)
Twenty-seven patients in the PTR arm had grade 3 or worse surgery-related adverse events, including anastomotic leakage in 3 and increased aspartate aminotransferase in 13. Three patients (4%) died within 30 days of surgery. Chemotherapy-related grade 3 or worse adverse events were more frequent and severe in the PTR arm (48% PTR versus 34% chemotherapy alone).
“For those who had been performing resections, this could push the paradigm back to chemotherapy alone,” Dr. Keller said.
Eleven patients (13%) in the chemotherapy-alone arm required surgery for intestinal obstruction or other primary tumor symptoms that developed after randomization, which means that 87% avoided surgery entirely, the investigators noted.
In the PTR group, surgery was performed within 21 days of enrollment, and chemotherapy started a median of 34 days after PTR. Chemotherapy was started within 14 days of enrollment in the chemotherapy-alone arm.
The groups were well balanced, including colon and rectosigmoid tumor locations in 93% of both arms and liver metastases in 73% of both. A bit over half the subjects were men and the median age was 65 years.
The team noted that early termination meant that the planned sample size was not achieved, which in turn limited the statistical power of the conclusions. “It is hoped that the comprehensive results of [ongoing trials] will clearly demonstrate the role of PTR for these patients,” they said.
The study was conducted by the Japan Clinical Oncology Group at 38 cancer centers in Japan. The work was funded by the Ministry of Health of Japan. The investigators had numerous industry ties, including Dr. Kanemitsu, who reported honoraria from Chugai Pharma, Ethicon, Covidien, and Intuitive Surgical, and being a Covidien adviser.
FROM THE JOURNAL OF CLINICAL ONCOLOGY