User login
Targeting CD98 to treat AML
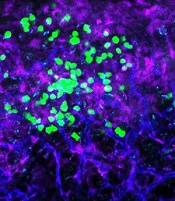
with blood vessels (blue).
Image courtesy of
UC San Diego Health
Preclinical research suggests the cell surface molecule CD98 promotes acute myeloid leukemia (AML), and the anti-CD98 antibody IGN523 can inhibit AML growth.
In AML patient cells and mouse models of the disease, IGN523 disrupted the interactions between leukemia cells and the surrounding blood vessels, thereby inhibiting the growth of AML.
Tannishtha Reya, PhD, of the University of California San Diego School of Medicine, and her colleagues reported these findings in Cancer Cell.
The team believes their results suggest IGN523 or other anti-CD98 antibodies might be useful for treating AML, particularly in children.
However, in a phase 1 study presented at the 2015 ASH Annual Meeting, IGN523 demonstrated only modest anti-leukemic activity in adults with AML.
Still, the researchers involved in the phase 1 study said IGN523 may prove effective in combination with other drugs used to treat AML.
Cancer Cell study
“To improve therapeutic strategies for [AML], we need to look not just at the cancer cells themselves but also at their interactions with surrounding cells, tissues, molecules, and blood vessels in the body,” Dr Reya said.
“In this study, we identified CD98 as a critical molecule driving AML growth. We showed that blocking CD98 can effectively reduce leukemia burden and improve survival by preventing cancer cells from receiving support from the surrounding environment.”
Dr Reya’s team engineered mouse models that lacked CD98 and found that loss of this molecule blocked AML growth and improved survival. Furthermore, CD98 loss largely spared normal blood cells, which the researchers said indicates a potential therapeutic window.
Additional experiments revealed that leukemia cells lacking CD98 had fewer stable interactions with the lining of blood vessels—interactions that were needed to fuel AML growth.
So the researchers decided to test the effects of blocking CD98 with a therapeutic inhibitor—IGN523. The team found that IGN523 blocks CD98’s AML-promoting activity in mouse models and human AML cells.
The researchers also transplanted patient-derived AML cells into mice and treated the recipients with either IGN523 or a control antibody. Anti-CD98 treatment effectively eliminated AML cells, while AML in the control mice expanded more than 100-fold.
“This study suggests that human AML can’t get established without CD98 and that blocking the molecule with anti-CD98 antibodies could be beneficial for the treatment of AML in both adults and children,” Dr Reya said.
Moving forward, Dr Reya and her colleagues are working to further define whether CD98 could be used to treat pediatric AML.
“Many of the models we used in this work were based on mutations found in childhood AML,” Dr Reya said. “While many childhood cancers have become very treatable, childhood AML continues to have a high rate of relapse and death.”
“We plan to work with pediatric oncologists to test if anti-CD98 agents can be effective against pediatric AML and whether it can improve responses to current treatments. I think this is particularly important to pursue since the anti-CD98 antibody has already been through phase 1 trials and could be more easily positioned to test in drug-resistant pediatric AML.”
Igenica Biotherapeutics Inc., the company developing IGN523, provided the drug for this study, and one of the study’s authors is an employee of the company. ![]()

with blood vessels (blue).
Image courtesy of
UC San Diego Health
Preclinical research suggests the cell surface molecule CD98 promotes acute myeloid leukemia (AML), and the anti-CD98 antibody IGN523 can inhibit AML growth.
In AML patient cells and mouse models of the disease, IGN523 disrupted the interactions between leukemia cells and the surrounding blood vessels, thereby inhibiting the growth of AML.
Tannishtha Reya, PhD, of the University of California San Diego School of Medicine, and her colleagues reported these findings in Cancer Cell.
The team believes their results suggest IGN523 or other anti-CD98 antibodies might be useful for treating AML, particularly in children.
However, in a phase 1 study presented at the 2015 ASH Annual Meeting, IGN523 demonstrated only modest anti-leukemic activity in adults with AML.
Still, the researchers involved in the phase 1 study said IGN523 may prove effective in combination with other drugs used to treat AML.
Cancer Cell study
“To improve therapeutic strategies for [AML], we need to look not just at the cancer cells themselves but also at their interactions with surrounding cells, tissues, molecules, and blood vessels in the body,” Dr Reya said.
“In this study, we identified CD98 as a critical molecule driving AML growth. We showed that blocking CD98 can effectively reduce leukemia burden and improve survival by preventing cancer cells from receiving support from the surrounding environment.”
Dr Reya’s team engineered mouse models that lacked CD98 and found that loss of this molecule blocked AML growth and improved survival. Furthermore, CD98 loss largely spared normal blood cells, which the researchers said indicates a potential therapeutic window.
Additional experiments revealed that leukemia cells lacking CD98 had fewer stable interactions with the lining of blood vessels—interactions that were needed to fuel AML growth.
So the researchers decided to test the effects of blocking CD98 with a therapeutic inhibitor—IGN523. The team found that IGN523 blocks CD98’s AML-promoting activity in mouse models and human AML cells.
The researchers also transplanted patient-derived AML cells into mice and treated the recipients with either IGN523 or a control antibody. Anti-CD98 treatment effectively eliminated AML cells, while AML in the control mice expanded more than 100-fold.
“This study suggests that human AML can’t get established without CD98 and that blocking the molecule with anti-CD98 antibodies could be beneficial for the treatment of AML in both adults and children,” Dr Reya said.
Moving forward, Dr Reya and her colleagues are working to further define whether CD98 could be used to treat pediatric AML.
“Many of the models we used in this work were based on mutations found in childhood AML,” Dr Reya said. “While many childhood cancers have become very treatable, childhood AML continues to have a high rate of relapse and death.”
“We plan to work with pediatric oncologists to test if anti-CD98 agents can be effective against pediatric AML and whether it can improve responses to current treatments. I think this is particularly important to pursue since the anti-CD98 antibody has already been through phase 1 trials and could be more easily positioned to test in drug-resistant pediatric AML.”
Igenica Biotherapeutics Inc., the company developing IGN523, provided the drug for this study, and one of the study’s authors is an employee of the company. ![]()

with blood vessels (blue).
Image courtesy of
UC San Diego Health
Preclinical research suggests the cell surface molecule CD98 promotes acute myeloid leukemia (AML), and the anti-CD98 antibody IGN523 can inhibit AML growth.
In AML patient cells and mouse models of the disease, IGN523 disrupted the interactions between leukemia cells and the surrounding blood vessels, thereby inhibiting the growth of AML.
Tannishtha Reya, PhD, of the University of California San Diego School of Medicine, and her colleagues reported these findings in Cancer Cell.
The team believes their results suggest IGN523 or other anti-CD98 antibodies might be useful for treating AML, particularly in children.
However, in a phase 1 study presented at the 2015 ASH Annual Meeting, IGN523 demonstrated only modest anti-leukemic activity in adults with AML.
Still, the researchers involved in the phase 1 study said IGN523 may prove effective in combination with other drugs used to treat AML.
Cancer Cell study
“To improve therapeutic strategies for [AML], we need to look not just at the cancer cells themselves but also at their interactions with surrounding cells, tissues, molecules, and blood vessels in the body,” Dr Reya said.
“In this study, we identified CD98 as a critical molecule driving AML growth. We showed that blocking CD98 can effectively reduce leukemia burden and improve survival by preventing cancer cells from receiving support from the surrounding environment.”
Dr Reya’s team engineered mouse models that lacked CD98 and found that loss of this molecule blocked AML growth and improved survival. Furthermore, CD98 loss largely spared normal blood cells, which the researchers said indicates a potential therapeutic window.
Additional experiments revealed that leukemia cells lacking CD98 had fewer stable interactions with the lining of blood vessels—interactions that were needed to fuel AML growth.
So the researchers decided to test the effects of blocking CD98 with a therapeutic inhibitor—IGN523. The team found that IGN523 blocks CD98’s AML-promoting activity in mouse models and human AML cells.
The researchers also transplanted patient-derived AML cells into mice and treated the recipients with either IGN523 or a control antibody. Anti-CD98 treatment effectively eliminated AML cells, while AML in the control mice expanded more than 100-fold.
“This study suggests that human AML can’t get established without CD98 and that blocking the molecule with anti-CD98 antibodies could be beneficial for the treatment of AML in both adults and children,” Dr Reya said.
Moving forward, Dr Reya and her colleagues are working to further define whether CD98 could be used to treat pediatric AML.
“Many of the models we used in this work were based on mutations found in childhood AML,” Dr Reya said. “While many childhood cancers have become very treatable, childhood AML continues to have a high rate of relapse and death.”
“We plan to work with pediatric oncologists to test if anti-CD98 agents can be effective against pediatric AML and whether it can improve responses to current treatments. I think this is particularly important to pursue since the anti-CD98 antibody has already been through phase 1 trials and could be more easily positioned to test in drug-resistant pediatric AML.”
Igenica Biotherapeutics Inc., the company developing IGN523, provided the drug for this study, and one of the study’s authors is an employee of the company. ![]()
Study reveals ‘high-traffic’ routes of malaria importation
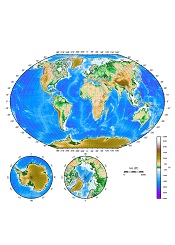
Results of an international study suggest France and the UK experience the highest number of malaria cases imported from other countries.
Researchers mapped the movement of malaria from endemic countries to 40 countries defined as being malaria-free.
The countries with the highest average number of imported infections per year over the past decade were France (2169), the UK (1898), the US (1511), Italy (637), and Germany (401).
Infection movement was strongly skewed to a small number of “high-traffic” routes, with malaria cases originating from West Africa accounting for 56% (13,947) of all cases detected in non-endemic countries.
These results were published in The Lancet Infectious Diseases.
“This is the first world-wide assessment of imported malaria cases in 20 years, and mapping this data is hugely valuable in helping us understand how we can mitigate against the effects of the global movements of the disease,” said study author Andrew Tatem, PhD, of the University of Southampton in the UK.
“Imported malaria can be expensive to treat, contribute to drug resistance, sometimes cause secondary local transmission, and threaten the long-term goal of eradication. This study forms part of wider efforts to understand patterns of human and malaria parasite movement to help guide elimination strategies.”
For this study, researchers analyzed a database of nationally reported statistics on imported malaria covering more than 50,000 individual cases over 10 years.
Although most incidents of malaria in non-endemic countries originated in West Africa, the study showed that 20% were from India (4988), 13% were from East Africa (3242), and 3% were from Papua New Guinea (748).
And although the routes from West Africa to France and the UK showed the strongest imported malaria link, there were other high-traffic routes. These included India to the US (149 cases on average per year), Papua New Guinea to Australia (97), Pakistan to the UK (69), and Haiti to the US (52).
By mapping this network of malaria movements across continents, the researchers showed that a number of factors, beyond geographic ones, may influence the strength of importation levels.
For example, the researchers believe that historical, economic, language, and cultural ties all play a part. They said population movements with former colonies had particular influence; such as Nigeria, Ghana, and Kenya with the UK, and Mali, Niger, and Chad with France.
The researchers hope to conduct further studies to examine which factors are the drivers behind the patterns of malaria spread between endemic and non-endemic countries. ![]()

Results of an international study suggest France and the UK experience the highest number of malaria cases imported from other countries.
Researchers mapped the movement of malaria from endemic countries to 40 countries defined as being malaria-free.
The countries with the highest average number of imported infections per year over the past decade were France (2169), the UK (1898), the US (1511), Italy (637), and Germany (401).
Infection movement was strongly skewed to a small number of “high-traffic” routes, with malaria cases originating from West Africa accounting for 56% (13,947) of all cases detected in non-endemic countries.
These results were published in The Lancet Infectious Diseases.
“This is the first world-wide assessment of imported malaria cases in 20 years, and mapping this data is hugely valuable in helping us understand how we can mitigate against the effects of the global movements of the disease,” said study author Andrew Tatem, PhD, of the University of Southampton in the UK.
“Imported malaria can be expensive to treat, contribute to drug resistance, sometimes cause secondary local transmission, and threaten the long-term goal of eradication. This study forms part of wider efforts to understand patterns of human and malaria parasite movement to help guide elimination strategies.”
For this study, researchers analyzed a database of nationally reported statistics on imported malaria covering more than 50,000 individual cases over 10 years.
Although most incidents of malaria in non-endemic countries originated in West Africa, the study showed that 20% were from India (4988), 13% were from East Africa (3242), and 3% were from Papua New Guinea (748).
And although the routes from West Africa to France and the UK showed the strongest imported malaria link, there were other high-traffic routes. These included India to the US (149 cases on average per year), Papua New Guinea to Australia (97), Pakistan to the UK (69), and Haiti to the US (52).
By mapping this network of malaria movements across continents, the researchers showed that a number of factors, beyond geographic ones, may influence the strength of importation levels.
For example, the researchers believe that historical, economic, language, and cultural ties all play a part. They said population movements with former colonies had particular influence; such as Nigeria, Ghana, and Kenya with the UK, and Mali, Niger, and Chad with France.
The researchers hope to conduct further studies to examine which factors are the drivers behind the patterns of malaria spread between endemic and non-endemic countries. ![]()

Results of an international study suggest France and the UK experience the highest number of malaria cases imported from other countries.
Researchers mapped the movement of malaria from endemic countries to 40 countries defined as being malaria-free.
The countries with the highest average number of imported infections per year over the past decade were France (2169), the UK (1898), the US (1511), Italy (637), and Germany (401).
Infection movement was strongly skewed to a small number of “high-traffic” routes, with malaria cases originating from West Africa accounting for 56% (13,947) of all cases detected in non-endemic countries.
These results were published in The Lancet Infectious Diseases.
“This is the first world-wide assessment of imported malaria cases in 20 years, and mapping this data is hugely valuable in helping us understand how we can mitigate against the effects of the global movements of the disease,” said study author Andrew Tatem, PhD, of the University of Southampton in the UK.
“Imported malaria can be expensive to treat, contribute to drug resistance, sometimes cause secondary local transmission, and threaten the long-term goal of eradication. This study forms part of wider efforts to understand patterns of human and malaria parasite movement to help guide elimination strategies.”
For this study, researchers analyzed a database of nationally reported statistics on imported malaria covering more than 50,000 individual cases over 10 years.
Although most incidents of malaria in non-endemic countries originated in West Africa, the study showed that 20% were from India (4988), 13% were from East Africa (3242), and 3% were from Papua New Guinea (748).
And although the routes from West Africa to France and the UK showed the strongest imported malaria link, there were other high-traffic routes. These included India to the US (149 cases on average per year), Papua New Guinea to Australia (97), Pakistan to the UK (69), and Haiti to the US (52).
By mapping this network of malaria movements across continents, the researchers showed that a number of factors, beyond geographic ones, may influence the strength of importation levels.
For example, the researchers believe that historical, economic, language, and cultural ties all play a part. They said population movements with former colonies had particular influence; such as Nigeria, Ghana, and Kenya with the UK, and Mali, Niger, and Chad with France.
The researchers hope to conduct further studies to examine which factors are the drivers behind the patterns of malaria spread between endemic and non-endemic countries. ![]()
Testing could ID cancer patients at high risk of VTE
Image by Andre E.X. Brown
Genetic testing could help identify breast cancer patients with a high risk of developing venous thromboembolism (VTE), according to a study published in Clinical Cancer Research.
The study showed that patients who received chemotherapy, had a higher genetic susceptibility for VTE according to a polygenic risk score (PRS), or had both of these risk factors were more likely to develop VTE than patients without these risk factors.
In addition, researchers found the impact of genetic susceptibility was most pronounced in older patients.
“The risk for [VTE] is increased in cancer patients, particularly in those receiving chemotherapy,” said study author Judith S. Brand, PhD, of Karolinska Institutet in Stockholm, Sweden.
“As one of the most common cancers, breast cancer accounts for a large number of cancer-associated VTE cases.”
Dr Brand and her colleagues sought to identify the individual and joint effects of chemotherapy and genetic susceptibility on VTE risk. They studied 4261 women in the Stockholm region diagnosed with primary invasive breast cancer between 2001 and 2008, and followed until 2012.
Risks were stratified based on chemotherapy status and genetic susceptibility, as determined by a PRS based on 9 established VTE loci, with the top 5% classified as having high genetic susceptibility.
The median follow-up was 7.6 years, and 276 patients experienced a VTE during that time.
The researchers found that receiving chemotherapy and having high genetic susceptibility independently increased the risk of VTE. The hazard ratio was 1.98 for patients receiving chemotherapy and 1.90 for patients in the highest 5% of the PRS.
The 1-year cumulative incidence of VTE was 9.5% among patients who both received chemotherapy and had high genetic susceptibility, compared with 1.3% in the patients who did not have either of these risk factors (P<0.001).
The researchers also found that patient age played a role in VTE risk. The team said the risk-increasing effect of the PRS was stronger in older patients (P interaction = 0.04).
In patients age 60 or older who underwent chemotherapy and had a high genetic susceptibility, the 1-year cumulative incidence of VTE was 25%.
“Breast cancer patients receiving chemotherapy are not routinely being examined for VTE prevention in today’s clinical practice,” Dr Brand said. “Our study demonstrates that information on genetic susceptibility can be used to identify patients at high risk of developing VTE.”
“Combined with other clinical risk factors and biomarkers, these findings will guide future studies evaluating routine VTE risk assessment in chemotherapy outpatients, and prophylaxis for those at highest risk. Because older patients demonstrated a stronger genetic effect and higher VTE incidence, this group requires special attention in future risk stratification efforts.”
Dr Brand added that a limitation of this study is the small number of older patients who had chemotherapy and a high genetic susceptibility. She said larger-scale studies would be necessary to provide more precise risk estimates. And further research is needed to assess the safety and potential benefit of thromboprophylaxis in high-risk cancer patients. ![]()

Image by Andre E.X. Brown
Genetic testing could help identify breast cancer patients with a high risk of developing venous thromboembolism (VTE), according to a study published in Clinical Cancer Research.
The study showed that patients who received chemotherapy, had a higher genetic susceptibility for VTE according to a polygenic risk score (PRS), or had both of these risk factors were more likely to develop VTE than patients without these risk factors.
In addition, researchers found the impact of genetic susceptibility was most pronounced in older patients.
“The risk for [VTE] is increased in cancer patients, particularly in those receiving chemotherapy,” said study author Judith S. Brand, PhD, of Karolinska Institutet in Stockholm, Sweden.
“As one of the most common cancers, breast cancer accounts for a large number of cancer-associated VTE cases.”
Dr Brand and her colleagues sought to identify the individual and joint effects of chemotherapy and genetic susceptibility on VTE risk. They studied 4261 women in the Stockholm region diagnosed with primary invasive breast cancer between 2001 and 2008, and followed until 2012.
Risks were stratified based on chemotherapy status and genetic susceptibility, as determined by a PRS based on 9 established VTE loci, with the top 5% classified as having high genetic susceptibility.
The median follow-up was 7.6 years, and 276 patients experienced a VTE during that time.
The researchers found that receiving chemotherapy and having high genetic susceptibility independently increased the risk of VTE. The hazard ratio was 1.98 for patients receiving chemotherapy and 1.90 for patients in the highest 5% of the PRS.
The 1-year cumulative incidence of VTE was 9.5% among patients who both received chemotherapy and had high genetic susceptibility, compared with 1.3% in the patients who did not have either of these risk factors (P<0.001).
The researchers also found that patient age played a role in VTE risk. The team said the risk-increasing effect of the PRS was stronger in older patients (P interaction = 0.04).
In patients age 60 or older who underwent chemotherapy and had a high genetic susceptibility, the 1-year cumulative incidence of VTE was 25%.
“Breast cancer patients receiving chemotherapy are not routinely being examined for VTE prevention in today’s clinical practice,” Dr Brand said. “Our study demonstrates that information on genetic susceptibility can be used to identify patients at high risk of developing VTE.”
“Combined with other clinical risk factors and biomarkers, these findings will guide future studies evaluating routine VTE risk assessment in chemotherapy outpatients, and prophylaxis for those at highest risk. Because older patients demonstrated a stronger genetic effect and higher VTE incidence, this group requires special attention in future risk stratification efforts.”
Dr Brand added that a limitation of this study is the small number of older patients who had chemotherapy and a high genetic susceptibility. She said larger-scale studies would be necessary to provide more precise risk estimates. And further research is needed to assess the safety and potential benefit of thromboprophylaxis in high-risk cancer patients. ![]()

Image by Andre E.X. Brown
Genetic testing could help identify breast cancer patients with a high risk of developing venous thromboembolism (VTE), according to a study published in Clinical Cancer Research.
The study showed that patients who received chemotherapy, had a higher genetic susceptibility for VTE according to a polygenic risk score (PRS), or had both of these risk factors were more likely to develop VTE than patients without these risk factors.
In addition, researchers found the impact of genetic susceptibility was most pronounced in older patients.
“The risk for [VTE] is increased in cancer patients, particularly in those receiving chemotherapy,” said study author Judith S. Brand, PhD, of Karolinska Institutet in Stockholm, Sweden.
“As one of the most common cancers, breast cancer accounts for a large number of cancer-associated VTE cases.”
Dr Brand and her colleagues sought to identify the individual and joint effects of chemotherapy and genetic susceptibility on VTE risk. They studied 4261 women in the Stockholm region diagnosed with primary invasive breast cancer between 2001 and 2008, and followed until 2012.
Risks were stratified based on chemotherapy status and genetic susceptibility, as determined by a PRS based on 9 established VTE loci, with the top 5% classified as having high genetic susceptibility.
The median follow-up was 7.6 years, and 276 patients experienced a VTE during that time.
The researchers found that receiving chemotherapy and having high genetic susceptibility independently increased the risk of VTE. The hazard ratio was 1.98 for patients receiving chemotherapy and 1.90 for patients in the highest 5% of the PRS.
The 1-year cumulative incidence of VTE was 9.5% among patients who both received chemotherapy and had high genetic susceptibility, compared with 1.3% in the patients who did not have either of these risk factors (P<0.001).
The researchers also found that patient age played a role in VTE risk. The team said the risk-increasing effect of the PRS was stronger in older patients (P interaction = 0.04).
In patients age 60 or older who underwent chemotherapy and had a high genetic susceptibility, the 1-year cumulative incidence of VTE was 25%.
“Breast cancer patients receiving chemotherapy are not routinely being examined for VTE prevention in today’s clinical practice,” Dr Brand said. “Our study demonstrates that information on genetic susceptibility can be used to identify patients at high risk of developing VTE.”
“Combined with other clinical risk factors and biomarkers, these findings will guide future studies evaluating routine VTE risk assessment in chemotherapy outpatients, and prophylaxis for those at highest risk. Because older patients demonstrated a stronger genetic effect and higher VTE incidence, this group requires special attention in future risk stratification efforts.”
Dr Brand added that a limitation of this study is the small number of older patients who had chemotherapy and a high genetic susceptibility. She said larger-scale studies would be necessary to provide more precise risk estimates. And further research is needed to assess the safety and potential benefit of thromboprophylaxis in high-risk cancer patients. ![]()
CTC analysis as good as BM biopsy in MM, team says

Photo by Juan D. Alfonso
Analysis of circulating tumor cells (CTCs) can provide at least as much genetic information about multiple myeloma (MM) as a bone marrow (BM) biopsy, according to a new study.
Researchers found evidence to suggest that analyzing CTCs isolated from the peripheral blood of MM patients could help physicians monitor disease progression over time, track the emergence of drug resistance, and tailor therapies to individual patients.
Jens Lohr, MD, PhD, of the Broad Institute of MIT and Harvard in Cambridge, Massachusetts, and colleagues conducted this research and detailed their findings in Science Translational Medicine.
The researchers said they devised a method that was “highly sensitive” in detecting and sequencing single CTCs, even from patients with low tumor burden.
Specifically, the team found they could isolate CTCs from, and detect mutations in, blood samples from an MM patient who had achieved a very good partial response to treatment and a patient with monoclonal gammopathy of undetermined significance.
Overall, the researchers found that CTCs could be used to identify mutations relevant for prognosis, and some of these mutations were more abundant in CTCs than in BM samples.
For example, the team detected somatic mutations in the KRAS, BRAF, IRF4, and TP53 genes in single CTCs from 3 MM patients. And the same mutations were present in single BM-derived MM cells.
In another 3 patients, the proportion of CTCs harboring TP53 R273C, BRAF G469A, and NRAS G13D mutations was higher than that observed in BM-derived cells. In 2 of the patients, the mutations weren’t detectable in BM cells because of insufficient sample material.
The researchers also found that CTCs could reveal patients with an overabundance of molecules expressed by MM cells—such as CD38 and SLAMF7—that can be targeted by therapies currently approved to treat MM—such as daratumumab and elotuzumab.
The team therefore believes that, with further development, CTC analysis could be a valuable tool for advancing precision medicine in MM. ![]()

Photo by Juan D. Alfonso
Analysis of circulating tumor cells (CTCs) can provide at least as much genetic information about multiple myeloma (MM) as a bone marrow (BM) biopsy, according to a new study.
Researchers found evidence to suggest that analyzing CTCs isolated from the peripheral blood of MM patients could help physicians monitor disease progression over time, track the emergence of drug resistance, and tailor therapies to individual patients.
Jens Lohr, MD, PhD, of the Broad Institute of MIT and Harvard in Cambridge, Massachusetts, and colleagues conducted this research and detailed their findings in Science Translational Medicine.
The researchers said they devised a method that was “highly sensitive” in detecting and sequencing single CTCs, even from patients with low tumor burden.
Specifically, the team found they could isolate CTCs from, and detect mutations in, blood samples from an MM patient who had achieved a very good partial response to treatment and a patient with monoclonal gammopathy of undetermined significance.
Overall, the researchers found that CTCs could be used to identify mutations relevant for prognosis, and some of these mutations were more abundant in CTCs than in BM samples.
For example, the team detected somatic mutations in the KRAS, BRAF, IRF4, and TP53 genes in single CTCs from 3 MM patients. And the same mutations were present in single BM-derived MM cells.
In another 3 patients, the proportion of CTCs harboring TP53 R273C, BRAF G469A, and NRAS G13D mutations was higher than that observed in BM-derived cells. In 2 of the patients, the mutations weren’t detectable in BM cells because of insufficient sample material.
The researchers also found that CTCs could reveal patients with an overabundance of molecules expressed by MM cells—such as CD38 and SLAMF7—that can be targeted by therapies currently approved to treat MM—such as daratumumab and elotuzumab.
The team therefore believes that, with further development, CTC analysis could be a valuable tool for advancing precision medicine in MM. ![]()

Photo by Juan D. Alfonso
Analysis of circulating tumor cells (CTCs) can provide at least as much genetic information about multiple myeloma (MM) as a bone marrow (BM) biopsy, according to a new study.
Researchers found evidence to suggest that analyzing CTCs isolated from the peripheral blood of MM patients could help physicians monitor disease progression over time, track the emergence of drug resistance, and tailor therapies to individual patients.
Jens Lohr, MD, PhD, of the Broad Institute of MIT and Harvard in Cambridge, Massachusetts, and colleagues conducted this research and detailed their findings in Science Translational Medicine.
The researchers said they devised a method that was “highly sensitive” in detecting and sequencing single CTCs, even from patients with low tumor burden.
Specifically, the team found they could isolate CTCs from, and detect mutations in, blood samples from an MM patient who had achieved a very good partial response to treatment and a patient with monoclonal gammopathy of undetermined significance.
Overall, the researchers found that CTCs could be used to identify mutations relevant for prognosis, and some of these mutations were more abundant in CTCs than in BM samples.
For example, the team detected somatic mutations in the KRAS, BRAF, IRF4, and TP53 genes in single CTCs from 3 MM patients. And the same mutations were present in single BM-derived MM cells.
In another 3 patients, the proportion of CTCs harboring TP53 R273C, BRAF G469A, and NRAS G13D mutations was higher than that observed in BM-derived cells. In 2 of the patients, the mutations weren’t detectable in BM cells because of insufficient sample material.
The researchers also found that CTCs could reveal patients with an overabundance of molecules expressed by MM cells—such as CD38 and SLAMF7—that can be targeted by therapies currently approved to treat MM—such as daratumumab and elotuzumab.
The team therefore believes that, with further development, CTC analysis could be a valuable tool for advancing precision medicine in MM. ![]()
Drug can fight adenovirus in HSCT recipients
Image by Yale Rosen
NEW ORLEANS—Interim results of a phase 3 trial suggest brincidofovir can treat adenovirus (AdV) infection in recipients of allogeneic hematopoietic stem cell transplant (HSCT).
Both pediatric and adult patients experienced a decline in AdV viral load after brincidofovir treatment, but pediatric patients were more likely to respond.
Overall survival rates were better for patients who had a rapid response and were therefore better among pediatric patients than adults.
Investigators said the adverse events (AEs) in this study were consistent with the known safety profile of brincidofovir.
Michael Grimley, MD, of Cincinnati Children’s Hospital in Ohio, and his colleagues presented these results at IDWeek 2016 (abstract 2339). The research was supported by Chimerix, the company developing brincidofovir.
This trial, known as AdVise, was designed to evaluate brincidofovir for the treatment of AdV infection in pediatric and adult patients divided into 3 cohorts:
- Cohort A consists of allogeneic HSCT recipients with asymptomatic or limited AdV infection
- Cohort B consists of allogeneic HSCT recipients with disseminated AdV disease
- Cohort C consists of autologous HSCT recipients, solid organ transplant recipients, and other immunocompromised patients.
All patients were assigned to 12 weeks of oral brincidofovir, administered twice weekly. An additional 12 weeks of treatment was allowed in patients with ongoing or recurrent infection. After completing treatment, all patients were followed until week 36.
Interim analysis
The investigators examined outcomes at 24 weeks after the first brincidofovir dose (12 weeks after prescribed dosing duration) in 158 patients, including:
- Cohort A—23 adults and 43 pediatric patients
- Cohort B—35 adults and 57 pediatric patients.
The investigators noted that many of the patients did not complete the study. The team said this is a reflection of the significant mortality risk of AdV because most of these patients died before they could finish.
Sixty-five percent of adults and 33% of children in Cohort A did not complete the study. The same was true for 71% of adults and 49% of children in Cohort B.
Mortality
The study’s primary efficacy endpoint is all-cause mortality at day 60 after the first brincidofovir dose in allogeneic HSCT recipients with disseminated AdV disease (Cohort B). All-cause mortality at day 60 in this cohort was 19% in pediatric patients and 43% in adults.
In Cohorts A and B, all-cause mortality at 24 weeks was lower in children than adults.
At 24 weeks, pediatric all-cause mortality was 33% in Cohort A and 42% in Cohort B. Adult all-cause mortality was 48% in Cohort A and 71% in Cohort B.
AdV-related mortality at 24 weeks in pediatric patients was 9% in Cohort A and 14% in Cohort B. AdV-related mortality in adults was 4% in Cohort A and 46% in Cohort B.
Declines in viremia
In Cohort A, 61% of patients achieved undetectable viremia at the end of treatment—43% of adults and 70% of children.
In Cohort B, 49% of patients achieved undetectable viremia at the end of treatment—29% of adults and 63% of children.
The median time to undetectable AdV viremia was 43 days (range, 8 to non-estimable) for adults in Cohort A, 14 days (range, 5 to 23) for children in Cohort A, non-estimable (range, 29 days to non-estimable) for adults in Cohort B, and 22 days (range, 15 to 36) for children in Cohort B.
Link between response and survival
The investigators conducted post-hoc analyses to assess the correlation between rapid virologic response to brincidofovir and time to subsequent mortality.
The team compared patients who responded to treatment—defined as achieving a ≥ 2-log10 copies/mL decline, undetectable AdV viremia at week 4, or undetectable AdV viremia at week 6—with non-responders.
Fifty percent of adults and 84% of children who were still alive at week 4 had achieved a ≥ 2 log decline or undetectable AdV viremia at that time.
This type of response was associated with improved survival at week 24. In adults, the mortality rate was 46% in responders and 85% in non-responders (P=0.03). In pediatric patients, the mortality rate was 25% in responders and 71% in non-responders (P=0.01).
In patients who were alive at week 6, 42% of adults and 68% of children achieved undetectable AdV viremia by that time.
This response was associated with improved survival at week 24. In adults, the mortality rate was 30% in responders and 86% in non-responders (P=0.001). In pediatric patients, the mortality rate was 18% in responders and 54% in non-responders (P=0.01).
Safety
All adults had treatment-emergent AEs, as did all pediatric patients in Cohort B and 95% of pediatric patients in Cohort A.
The most common treatment-emergent AEs were gastrointestinal (GI) events, which occurred in 70% of adults and 81% of children in Cohort A, as well as 83% of adults and 74% of children in Cohort B.
Acute graft-versus-host disease (GVHD) was also common, occurring in 22% of adults and 37% of children in Cohort A and 43% of adults and 40% of children in Cohort B. Some patients did have acute GVHD at baseline, however—22%, 26%, 34%, and 19%, respectively.
The percentage of patients with AEs leading to treatment discontinuation was 26% for adults and 28% for children in Cohort A and 31% for adults and 14% for children in Cohort B.
Overall, 20% of pediatric patients and 29% of adults discontinued brincidofovir due to AEs. GI events were cited as the most common reason—5% and 14%, respectively.
The investigators said there were no events reported that were suggestive of drug-related nephrotoxicity or myelosuppression. ![]()

Image by Yale Rosen
NEW ORLEANS—Interim results of a phase 3 trial suggest brincidofovir can treat adenovirus (AdV) infection in recipients of allogeneic hematopoietic stem cell transplant (HSCT).
Both pediatric and adult patients experienced a decline in AdV viral load after brincidofovir treatment, but pediatric patients were more likely to respond.
Overall survival rates were better for patients who had a rapid response and were therefore better among pediatric patients than adults.
Investigators said the adverse events (AEs) in this study were consistent with the known safety profile of brincidofovir.
Michael Grimley, MD, of Cincinnati Children’s Hospital in Ohio, and his colleagues presented these results at IDWeek 2016 (abstract 2339). The research was supported by Chimerix, the company developing brincidofovir.
This trial, known as AdVise, was designed to evaluate brincidofovir for the treatment of AdV infection in pediatric and adult patients divided into 3 cohorts:
- Cohort A consists of allogeneic HSCT recipients with asymptomatic or limited AdV infection
- Cohort B consists of allogeneic HSCT recipients with disseminated AdV disease
- Cohort C consists of autologous HSCT recipients, solid organ transplant recipients, and other immunocompromised patients.
All patients were assigned to 12 weeks of oral brincidofovir, administered twice weekly. An additional 12 weeks of treatment was allowed in patients with ongoing or recurrent infection. After completing treatment, all patients were followed until week 36.
Interim analysis
The investigators examined outcomes at 24 weeks after the first brincidofovir dose (12 weeks after prescribed dosing duration) in 158 patients, including:
- Cohort A—23 adults and 43 pediatric patients
- Cohort B—35 adults and 57 pediatric patients.
The investigators noted that many of the patients did not complete the study. The team said this is a reflection of the significant mortality risk of AdV because most of these patients died before they could finish.
Sixty-five percent of adults and 33% of children in Cohort A did not complete the study. The same was true for 71% of adults and 49% of children in Cohort B.
Mortality
The study’s primary efficacy endpoint is all-cause mortality at day 60 after the first brincidofovir dose in allogeneic HSCT recipients with disseminated AdV disease (Cohort B). All-cause mortality at day 60 in this cohort was 19% in pediatric patients and 43% in adults.
In Cohorts A and B, all-cause mortality at 24 weeks was lower in children than adults.
At 24 weeks, pediatric all-cause mortality was 33% in Cohort A and 42% in Cohort B. Adult all-cause mortality was 48% in Cohort A and 71% in Cohort B.
AdV-related mortality at 24 weeks in pediatric patients was 9% in Cohort A and 14% in Cohort B. AdV-related mortality in adults was 4% in Cohort A and 46% in Cohort B.
Declines in viremia
In Cohort A, 61% of patients achieved undetectable viremia at the end of treatment—43% of adults and 70% of children.
In Cohort B, 49% of patients achieved undetectable viremia at the end of treatment—29% of adults and 63% of children.
The median time to undetectable AdV viremia was 43 days (range, 8 to non-estimable) for adults in Cohort A, 14 days (range, 5 to 23) for children in Cohort A, non-estimable (range, 29 days to non-estimable) for adults in Cohort B, and 22 days (range, 15 to 36) for children in Cohort B.
Link between response and survival
The investigators conducted post-hoc analyses to assess the correlation between rapid virologic response to brincidofovir and time to subsequent mortality.
The team compared patients who responded to treatment—defined as achieving a ≥ 2-log10 copies/mL decline, undetectable AdV viremia at week 4, or undetectable AdV viremia at week 6—with non-responders.
Fifty percent of adults and 84% of children who were still alive at week 4 had achieved a ≥ 2 log decline or undetectable AdV viremia at that time.
This type of response was associated with improved survival at week 24. In adults, the mortality rate was 46% in responders and 85% in non-responders (P=0.03). In pediatric patients, the mortality rate was 25% in responders and 71% in non-responders (P=0.01).
In patients who were alive at week 6, 42% of adults and 68% of children achieved undetectable AdV viremia by that time.
This response was associated with improved survival at week 24. In adults, the mortality rate was 30% in responders and 86% in non-responders (P=0.001). In pediatric patients, the mortality rate was 18% in responders and 54% in non-responders (P=0.01).
Safety
All adults had treatment-emergent AEs, as did all pediatric patients in Cohort B and 95% of pediatric patients in Cohort A.
The most common treatment-emergent AEs were gastrointestinal (GI) events, which occurred in 70% of adults and 81% of children in Cohort A, as well as 83% of adults and 74% of children in Cohort B.
Acute graft-versus-host disease (GVHD) was also common, occurring in 22% of adults and 37% of children in Cohort A and 43% of adults and 40% of children in Cohort B. Some patients did have acute GVHD at baseline, however—22%, 26%, 34%, and 19%, respectively.
The percentage of patients with AEs leading to treatment discontinuation was 26% for adults and 28% for children in Cohort A and 31% for adults and 14% for children in Cohort B.
Overall, 20% of pediatric patients and 29% of adults discontinued brincidofovir due to AEs. GI events were cited as the most common reason—5% and 14%, respectively.
The investigators said there were no events reported that were suggestive of drug-related nephrotoxicity or myelosuppression. ![]()

Image by Yale Rosen
NEW ORLEANS—Interim results of a phase 3 trial suggest brincidofovir can treat adenovirus (AdV) infection in recipients of allogeneic hematopoietic stem cell transplant (HSCT).
Both pediatric and adult patients experienced a decline in AdV viral load after brincidofovir treatment, but pediatric patients were more likely to respond.
Overall survival rates were better for patients who had a rapid response and were therefore better among pediatric patients than adults.
Investigators said the adverse events (AEs) in this study were consistent with the known safety profile of brincidofovir.
Michael Grimley, MD, of Cincinnati Children’s Hospital in Ohio, and his colleagues presented these results at IDWeek 2016 (abstract 2339). The research was supported by Chimerix, the company developing brincidofovir.
This trial, known as AdVise, was designed to evaluate brincidofovir for the treatment of AdV infection in pediatric and adult patients divided into 3 cohorts:
- Cohort A consists of allogeneic HSCT recipients with asymptomatic or limited AdV infection
- Cohort B consists of allogeneic HSCT recipients with disseminated AdV disease
- Cohort C consists of autologous HSCT recipients, solid organ transplant recipients, and other immunocompromised patients.
All patients were assigned to 12 weeks of oral brincidofovir, administered twice weekly. An additional 12 weeks of treatment was allowed in patients with ongoing or recurrent infection. After completing treatment, all patients were followed until week 36.
Interim analysis
The investigators examined outcomes at 24 weeks after the first brincidofovir dose (12 weeks after prescribed dosing duration) in 158 patients, including:
- Cohort A—23 adults and 43 pediatric patients
- Cohort B—35 adults and 57 pediatric patients.
The investigators noted that many of the patients did not complete the study. The team said this is a reflection of the significant mortality risk of AdV because most of these patients died before they could finish.
Sixty-five percent of adults and 33% of children in Cohort A did not complete the study. The same was true for 71% of adults and 49% of children in Cohort B.
Mortality
The study’s primary efficacy endpoint is all-cause mortality at day 60 after the first brincidofovir dose in allogeneic HSCT recipients with disseminated AdV disease (Cohort B). All-cause mortality at day 60 in this cohort was 19% in pediatric patients and 43% in adults.
In Cohorts A and B, all-cause mortality at 24 weeks was lower in children than adults.
At 24 weeks, pediatric all-cause mortality was 33% in Cohort A and 42% in Cohort B. Adult all-cause mortality was 48% in Cohort A and 71% in Cohort B.
AdV-related mortality at 24 weeks in pediatric patients was 9% in Cohort A and 14% in Cohort B. AdV-related mortality in adults was 4% in Cohort A and 46% in Cohort B.
Declines in viremia
In Cohort A, 61% of patients achieved undetectable viremia at the end of treatment—43% of adults and 70% of children.
In Cohort B, 49% of patients achieved undetectable viremia at the end of treatment—29% of adults and 63% of children.
The median time to undetectable AdV viremia was 43 days (range, 8 to non-estimable) for adults in Cohort A, 14 days (range, 5 to 23) for children in Cohort A, non-estimable (range, 29 days to non-estimable) for adults in Cohort B, and 22 days (range, 15 to 36) for children in Cohort B.
Link between response and survival
The investigators conducted post-hoc analyses to assess the correlation between rapid virologic response to brincidofovir and time to subsequent mortality.
The team compared patients who responded to treatment—defined as achieving a ≥ 2-log10 copies/mL decline, undetectable AdV viremia at week 4, or undetectable AdV viremia at week 6—with non-responders.
Fifty percent of adults and 84% of children who were still alive at week 4 had achieved a ≥ 2 log decline or undetectable AdV viremia at that time.
This type of response was associated with improved survival at week 24. In adults, the mortality rate was 46% in responders and 85% in non-responders (P=0.03). In pediatric patients, the mortality rate was 25% in responders and 71% in non-responders (P=0.01).
In patients who were alive at week 6, 42% of adults and 68% of children achieved undetectable AdV viremia by that time.
This response was associated with improved survival at week 24. In adults, the mortality rate was 30% in responders and 86% in non-responders (P=0.001). In pediatric patients, the mortality rate was 18% in responders and 54% in non-responders (P=0.01).
Safety
All adults had treatment-emergent AEs, as did all pediatric patients in Cohort B and 95% of pediatric patients in Cohort A.
The most common treatment-emergent AEs were gastrointestinal (GI) events, which occurred in 70% of adults and 81% of children in Cohort A, as well as 83% of adults and 74% of children in Cohort B.
Acute graft-versus-host disease (GVHD) was also common, occurring in 22% of adults and 37% of children in Cohort A and 43% of adults and 40% of children in Cohort B. Some patients did have acute GVHD at baseline, however—22%, 26%, 34%, and 19%, respectively.
The percentage of patients with AEs leading to treatment discontinuation was 26% for adults and 28% for children in Cohort A and 31% for adults and 14% for children in Cohort B.
Overall, 20% of pediatric patients and 29% of adults discontinued brincidofovir due to AEs. GI events were cited as the most common reason—5% and 14%, respectively.
The investigators said there were no events reported that were suggestive of drug-related nephrotoxicity or myelosuppression. ![]()
Two-drug combination targets LSCs in CML
Image by Difu Wu
Targeting a pair of transcription factors might improve the treatment of chronic myeloid leukemia (CML), according to researchers.
The team found that p53 and c-MYC have “defining roles” in the survival of leukemia stem cells (LSCs) in CML.
And by targeting these transcription factors with a pair of investigational drugs, the researchers were able to kill LSCs.
The team described this work in Nature.
“This collaborative study combined proteomics, transcriptomics, and systems biology to identify a novel, precision medicine-based approach for eradicating leukemic stem cells,” said study author Tony Whetton, PhD, of the University of Manchester in the UK.
Dr Whetton and his colleagues first discovered that p53 and c-MYC are “central hubs” in a CML network of deregulated proteins. The team also found that CML cells express increased c-MYC and decreased p53 levels.
So the researchers theorized that simultaneously activating p53 and inhibiting c-MYC could be a method for treating CML.
To that end, the team tested 2 drugs—RITA (or NSC652287), which binds p53 and blocks its degradation, and CPI-203, a BET inhibitor that hinders transcription by disrupting chromatin-dependent signal transduction.
The researchers found that CPI-203 successfully downregulated c-MYC but also reduced p53, while RITA increased p53.
Treating CML CD34+ cells with RITA or CPI-203 for 72 hours reduced cell viability and induced significant apoptosis, the team said. Combining the drugs enhanced these effects.
The researchers also found evidence to suggest that c-MYC inhibition induces differentiation of CML CD34+ cells. The team said that labelling with the cell-division tracker carboxyfluorescein succinimidyl ester (CFSE) and CD34 antibody showed that, as CML cells divided in the presence of CPI-203, there was a clear and rapid loss of CD34 expression that was not seen in the presence of RITA.
The researchers did not observe any differences in the effects of RITA and CPI-203 when they were tested in CML CD34+ cells pretreated with imatinib.
Furthermore, RITA and CPI-203, either alone or in combination, had no significant effects on normal CD34+ cells when tested at lower concentrations. However, when CPI-203 was used alone at higher concentrations (2 or 5 μ M) or with RITA at the highest concentrations tested (RITA at 25 nM, CPI-203 at 5 μ M), apoptosis did occur.
In CML cells, the researchers observed “significant apoptosis” with all concentrations of CPI-203 and RITA tested.
The team also exposed CML LSCs, defined as either CFSEmax or CD34+CD38− cells, to CPI-203 and RITA as well as a pair of tyrosine kinase inhibitors.
The CFSEmax population persisted despite 5 days of treatment with dasatinib or nilotinib, but the cells were “significantly reduced” after 5 days of treatment with CPI-203 alone and in combination with RITA.
Similarly, 72 hours of treatment with RITA with CPI-203 eliminated residual CD34+CD38− cells.
The researchers also assessed LSC engraftment after treatment with RITA and/or CPI-203, as well as dasatinib. They exposed CML CD34+ cells to the drugs for 48 hours before transplanting the cells into sublethally irradiated NSG mice.
The team said dasatinib had no significant effect on NSG-repopulating CML LSCs. However, RITA, CPI-203, and the drugs in combination reduced engraftment, as indicated by decreased CD45+, CD34+, CD33+, CD11b+, CD19+ and CD14+ cells. ![]()

Image by Difu Wu
Targeting a pair of transcription factors might improve the treatment of chronic myeloid leukemia (CML), according to researchers.
The team found that p53 and c-MYC have “defining roles” in the survival of leukemia stem cells (LSCs) in CML.
And by targeting these transcription factors with a pair of investigational drugs, the researchers were able to kill LSCs.
The team described this work in Nature.
“This collaborative study combined proteomics, transcriptomics, and systems biology to identify a novel, precision medicine-based approach for eradicating leukemic stem cells,” said study author Tony Whetton, PhD, of the University of Manchester in the UK.
Dr Whetton and his colleagues first discovered that p53 and c-MYC are “central hubs” in a CML network of deregulated proteins. The team also found that CML cells express increased c-MYC and decreased p53 levels.
So the researchers theorized that simultaneously activating p53 and inhibiting c-MYC could be a method for treating CML.
To that end, the team tested 2 drugs—RITA (or NSC652287), which binds p53 and blocks its degradation, and CPI-203, a BET inhibitor that hinders transcription by disrupting chromatin-dependent signal transduction.
The researchers found that CPI-203 successfully downregulated c-MYC but also reduced p53, while RITA increased p53.
Treating CML CD34+ cells with RITA or CPI-203 for 72 hours reduced cell viability and induced significant apoptosis, the team said. Combining the drugs enhanced these effects.
The researchers also found evidence to suggest that c-MYC inhibition induces differentiation of CML CD34+ cells. The team said that labelling with the cell-division tracker carboxyfluorescein succinimidyl ester (CFSE) and CD34 antibody showed that, as CML cells divided in the presence of CPI-203, there was a clear and rapid loss of CD34 expression that was not seen in the presence of RITA.
The researchers did not observe any differences in the effects of RITA and CPI-203 when they were tested in CML CD34+ cells pretreated with imatinib.
Furthermore, RITA and CPI-203, either alone or in combination, had no significant effects on normal CD34+ cells when tested at lower concentrations. However, when CPI-203 was used alone at higher concentrations (2 or 5 μ M) or with RITA at the highest concentrations tested (RITA at 25 nM, CPI-203 at 5 μ M), apoptosis did occur.
In CML cells, the researchers observed “significant apoptosis” with all concentrations of CPI-203 and RITA tested.
The team also exposed CML LSCs, defined as either CFSEmax or CD34+CD38− cells, to CPI-203 and RITA as well as a pair of tyrosine kinase inhibitors.
The CFSEmax population persisted despite 5 days of treatment with dasatinib or nilotinib, but the cells were “significantly reduced” after 5 days of treatment with CPI-203 alone and in combination with RITA.
Similarly, 72 hours of treatment with RITA with CPI-203 eliminated residual CD34+CD38− cells.
The researchers also assessed LSC engraftment after treatment with RITA and/or CPI-203, as well as dasatinib. They exposed CML CD34+ cells to the drugs for 48 hours before transplanting the cells into sublethally irradiated NSG mice.
The team said dasatinib had no significant effect on NSG-repopulating CML LSCs. However, RITA, CPI-203, and the drugs in combination reduced engraftment, as indicated by decreased CD45+, CD34+, CD33+, CD11b+, CD19+ and CD14+ cells. ![]()

Image by Difu Wu
Targeting a pair of transcription factors might improve the treatment of chronic myeloid leukemia (CML), according to researchers.
The team found that p53 and c-MYC have “defining roles” in the survival of leukemia stem cells (LSCs) in CML.
And by targeting these transcription factors with a pair of investigational drugs, the researchers were able to kill LSCs.
The team described this work in Nature.
“This collaborative study combined proteomics, transcriptomics, and systems biology to identify a novel, precision medicine-based approach for eradicating leukemic stem cells,” said study author Tony Whetton, PhD, of the University of Manchester in the UK.
Dr Whetton and his colleagues first discovered that p53 and c-MYC are “central hubs” in a CML network of deregulated proteins. The team also found that CML cells express increased c-MYC and decreased p53 levels.
So the researchers theorized that simultaneously activating p53 and inhibiting c-MYC could be a method for treating CML.
To that end, the team tested 2 drugs—RITA (or NSC652287), which binds p53 and blocks its degradation, and CPI-203, a BET inhibitor that hinders transcription by disrupting chromatin-dependent signal transduction.
The researchers found that CPI-203 successfully downregulated c-MYC but also reduced p53, while RITA increased p53.
Treating CML CD34+ cells with RITA or CPI-203 for 72 hours reduced cell viability and induced significant apoptosis, the team said. Combining the drugs enhanced these effects.
The researchers also found evidence to suggest that c-MYC inhibition induces differentiation of CML CD34+ cells. The team said that labelling with the cell-division tracker carboxyfluorescein succinimidyl ester (CFSE) and CD34 antibody showed that, as CML cells divided in the presence of CPI-203, there was a clear and rapid loss of CD34 expression that was not seen in the presence of RITA.
The researchers did not observe any differences in the effects of RITA and CPI-203 when they were tested in CML CD34+ cells pretreated with imatinib.
Furthermore, RITA and CPI-203, either alone or in combination, had no significant effects on normal CD34+ cells when tested at lower concentrations. However, when CPI-203 was used alone at higher concentrations (2 or 5 μ M) or with RITA at the highest concentrations tested (RITA at 25 nM, CPI-203 at 5 μ M), apoptosis did occur.
In CML cells, the researchers observed “significant apoptosis” with all concentrations of CPI-203 and RITA tested.
The team also exposed CML LSCs, defined as either CFSEmax or CD34+CD38− cells, to CPI-203 and RITA as well as a pair of tyrosine kinase inhibitors.
The CFSEmax population persisted despite 5 days of treatment with dasatinib or nilotinib, but the cells were “significantly reduced” after 5 days of treatment with CPI-203 alone and in combination with RITA.
Similarly, 72 hours of treatment with RITA with CPI-203 eliminated residual CD34+CD38− cells.
The researchers also assessed LSC engraftment after treatment with RITA and/or CPI-203, as well as dasatinib. They exposed CML CD34+ cells to the drugs for 48 hours before transplanting the cells into sublethally irradiated NSG mice.
The team said dasatinib had no significant effect on NSG-repopulating CML LSCs. However, RITA, CPI-203, and the drugs in combination reduced engraftment, as indicated by decreased CD45+, CD34+, CD33+, CD11b+, CD19+ and CD14+ cells. ![]()
Tranexamic acid safely reduces need for transfusion, study suggests
Photo by Piotr Bodzek
Results of a large study suggest that tranexamic acid can reduce the need for blood transfusion without increasing the risk of thrombotic complications or death in patients undergoing coronary artery surgery.
Patients who received tranexamic acid had a lower risk of excessive bleeding, required fewer units of blood products, and had a lower risk of emergency reoperation after surgery than patients who received placebo.
In addition, patients who received tranexamic acid had no higher risk of death or thrombotic complications than those who received placebo.
Paul S. Myles, MBBS, MD, of Alfred Hospital in Melbourne, Australia, and his colleagues conducted this study and reported the results in NEJM. The study was also presented at the ANESTHESIOLOGY® 2016 annual meeting.
The study included 4631 patients who underwent surgery and had available outcomes data, 2311 who were assigned to receive tranexamic acid and 2320 who were assigned to receive placebo.
The study’s primary outcome was a composite of death and thrombotic complications (nonfatal myocardial infarction, stroke, pulmonary embolism, renal failure, or bowel infarction) within 30 days after surgery.
There was no significant difference in the primary outcome between the 2 treatment groups. Thrombotic complications/death occurred in 16.7% of patients in the tranexamic acid group and 18.1% in the placebo group (relative risk=0.92; P=0.22).
Patients who received placebo required significantly more units of blood products than patients who received tranexamic acid—7994 and 4331 units, respectively (P<0.001).
And significantly fewer patients in the tranexamic acid group than the placebo group had major hemorrhage or cardiac tamponade leading to emergency reoperations—1.4% and 2.8%, respectively (P=0.001).
However, patients in the tranexamic group had a significantly higher incidence of seizures—0.7% and 0.1%, respectively (P=0.002).
Dr Myles said that although this study was conducted in patients undergoing coronary artery surgery, the results are relevant for patients having many other types of surgery where bleeding and the need for blood transfusion may occur. ![]()

Photo by Piotr Bodzek
Results of a large study suggest that tranexamic acid can reduce the need for blood transfusion without increasing the risk of thrombotic complications or death in patients undergoing coronary artery surgery.
Patients who received tranexamic acid had a lower risk of excessive bleeding, required fewer units of blood products, and had a lower risk of emergency reoperation after surgery than patients who received placebo.
In addition, patients who received tranexamic acid had no higher risk of death or thrombotic complications than those who received placebo.
Paul S. Myles, MBBS, MD, of Alfred Hospital in Melbourne, Australia, and his colleagues conducted this study and reported the results in NEJM. The study was also presented at the ANESTHESIOLOGY® 2016 annual meeting.
The study included 4631 patients who underwent surgery and had available outcomes data, 2311 who were assigned to receive tranexamic acid and 2320 who were assigned to receive placebo.
The study’s primary outcome was a composite of death and thrombotic complications (nonfatal myocardial infarction, stroke, pulmonary embolism, renal failure, or bowel infarction) within 30 days after surgery.
There was no significant difference in the primary outcome between the 2 treatment groups. Thrombotic complications/death occurred in 16.7% of patients in the tranexamic acid group and 18.1% in the placebo group (relative risk=0.92; P=0.22).
Patients who received placebo required significantly more units of blood products than patients who received tranexamic acid—7994 and 4331 units, respectively (P<0.001).
And significantly fewer patients in the tranexamic acid group than the placebo group had major hemorrhage or cardiac tamponade leading to emergency reoperations—1.4% and 2.8%, respectively (P=0.001).
However, patients in the tranexamic group had a significantly higher incidence of seizures—0.7% and 0.1%, respectively (P=0.002).
Dr Myles said that although this study was conducted in patients undergoing coronary artery surgery, the results are relevant for patients having many other types of surgery where bleeding and the need for blood transfusion may occur. ![]()

Photo by Piotr Bodzek
Results of a large study suggest that tranexamic acid can reduce the need for blood transfusion without increasing the risk of thrombotic complications or death in patients undergoing coronary artery surgery.
Patients who received tranexamic acid had a lower risk of excessive bleeding, required fewer units of blood products, and had a lower risk of emergency reoperation after surgery than patients who received placebo.
In addition, patients who received tranexamic acid had no higher risk of death or thrombotic complications than those who received placebo.
Paul S. Myles, MBBS, MD, of Alfred Hospital in Melbourne, Australia, and his colleagues conducted this study and reported the results in NEJM. The study was also presented at the ANESTHESIOLOGY® 2016 annual meeting.
The study included 4631 patients who underwent surgery and had available outcomes data, 2311 who were assigned to receive tranexamic acid and 2320 who were assigned to receive placebo.
The study’s primary outcome was a composite of death and thrombotic complications (nonfatal myocardial infarction, stroke, pulmonary embolism, renal failure, or bowel infarction) within 30 days after surgery.
There was no significant difference in the primary outcome between the 2 treatment groups. Thrombotic complications/death occurred in 16.7% of patients in the tranexamic acid group and 18.1% in the placebo group (relative risk=0.92; P=0.22).
Patients who received placebo required significantly more units of blood products than patients who received tranexamic acid—7994 and 4331 units, respectively (P<0.001).
And significantly fewer patients in the tranexamic acid group than the placebo group had major hemorrhage or cardiac tamponade leading to emergency reoperations—1.4% and 2.8%, respectively (P=0.001).
However, patients in the tranexamic group had a significantly higher incidence of seizures—0.7% and 0.1%, respectively (P=0.002).
Dr Myles said that although this study was conducted in patients undergoing coronary artery surgery, the results are relevant for patients having many other types of surgery where bleeding and the need for blood transfusion may occur.
Tobacco plants used to manufacture malaria drug

Tobacco plants can be engineered to manufacture artemisinin at therapeutic levels, according research published in Molecular Plant.
The researchers noted that the majority of people who live in malaria-endemic areas cannot afford to buy artemisinin.
The drug’s high cost is due to the extraction process and the fact that it’s difficult to grow Artemisia annua, the original source of the drug, in climates where malaria is common.
Advances in synthetic biology have made it possible to produce artemisinin in yeast, but the manufacturing process is difficult to scale up.
Earlier studies showed that artemisinin can be grown in tobacco—a plant that’s relatively easy to genetically manipulate and that grows well in areas where malaria is endemic. But yields of artemisinin from those plants were low.
Now, Shashi Kumar, PhD, of the International Centre for Genetic
Engineering and Biotechnology in New Delhi, India, and his colleagues say they have overcome this problem.
In the Molecular Plant paper, Dr Kumar and his colleagues reported using a dual-transformation approach to boost the production of artemisinin in the tobacco plants.
The team first generated plants that contained transgenic chloroplasts, and the same plants were then manipulated again to insert genes into the nuclear genome as well.
Extract from the plants was shown to stop the growth of Plasmodium falciparum in vitro. Whole cells from the plant were also fed to mice infected with Plasmodium berghei, which greatly reduced levels of the parasite in the blood.
In fact, the researchers found the whole plant material was more effective in attacking the parasite than pure artemisinin, likely because encapsulation inside the plant cells protected the compound from degradation by digestive enzymes.
The researchers acknowledged that convincing people to eat tobacco plants is likely to be a hard sell. For that reason, they are now aiming to genetically engineer lettuce plants to produce artemisinin at therapeutic levels.
They said the lettuce containing the drug could be freeze dried, ground into a powder, and put into capsules for cost-effective delivery.
“Plant and animal science are increasingly coming together,” Dr Kumar said. “In the near future, you will see more drugs produced inside plants will be commercialized to reduce the drug cost.”

Tobacco plants can be engineered to manufacture artemisinin at therapeutic levels, according research published in Molecular Plant.
The researchers noted that the majority of people who live in malaria-endemic areas cannot afford to buy artemisinin.
The drug’s high cost is due to the extraction process and the fact that it’s difficult to grow Artemisia annua, the original source of the drug, in climates where malaria is common.
Advances in synthetic biology have made it possible to produce artemisinin in yeast, but the manufacturing process is difficult to scale up.
Earlier studies showed that artemisinin can be grown in tobacco—a plant that’s relatively easy to genetically manipulate and that grows well in areas where malaria is endemic. But yields of artemisinin from those plants were low.
Now, Shashi Kumar, PhD, of the International Centre for Genetic
Engineering and Biotechnology in New Delhi, India, and his colleagues say they have overcome this problem.
In the Molecular Plant paper, Dr Kumar and his colleagues reported using a dual-transformation approach to boost the production of artemisinin in the tobacco plants.
The team first generated plants that contained transgenic chloroplasts, and the same plants were then manipulated again to insert genes into the nuclear genome as well.
Extract from the plants was shown to stop the growth of Plasmodium falciparum in vitro. Whole cells from the plant were also fed to mice infected with Plasmodium berghei, which greatly reduced levels of the parasite in the blood.
In fact, the researchers found the whole plant material was more effective in attacking the parasite than pure artemisinin, likely because encapsulation inside the plant cells protected the compound from degradation by digestive enzymes.
The researchers acknowledged that convincing people to eat tobacco plants is likely to be a hard sell. For that reason, they are now aiming to genetically engineer lettuce plants to produce artemisinin at therapeutic levels.
They said the lettuce containing the drug could be freeze dried, ground into a powder, and put into capsules for cost-effective delivery.
“Plant and animal science are increasingly coming together,” Dr Kumar said. “In the near future, you will see more drugs produced inside plants will be commercialized to reduce the drug cost.”

Tobacco plants can be engineered to manufacture artemisinin at therapeutic levels, according research published in Molecular Plant.
The researchers noted that the majority of people who live in malaria-endemic areas cannot afford to buy artemisinin.
The drug’s high cost is due to the extraction process and the fact that it’s difficult to grow Artemisia annua, the original source of the drug, in climates where malaria is common.
Advances in synthetic biology have made it possible to produce artemisinin in yeast, but the manufacturing process is difficult to scale up.
Earlier studies showed that artemisinin can be grown in tobacco—a plant that’s relatively easy to genetically manipulate and that grows well in areas where malaria is endemic. But yields of artemisinin from those plants were low.
Now, Shashi Kumar, PhD, of the International Centre for Genetic
Engineering and Biotechnology in New Delhi, India, and his colleagues say they have overcome this problem.
In the Molecular Plant paper, Dr Kumar and his colleagues reported using a dual-transformation approach to boost the production of artemisinin in the tobacco plants.
The team first generated plants that contained transgenic chloroplasts, and the same plants were then manipulated again to insert genes into the nuclear genome as well.
Extract from the plants was shown to stop the growth of Plasmodium falciparum in vitro. Whole cells from the plant were also fed to mice infected with Plasmodium berghei, which greatly reduced levels of the parasite in the blood.
In fact, the researchers found the whole plant material was more effective in attacking the parasite than pure artemisinin, likely because encapsulation inside the plant cells protected the compound from degradation by digestive enzymes.
The researchers acknowledged that convincing people to eat tobacco plants is likely to be a hard sell. For that reason, they are now aiming to genetically engineer lettuce plants to produce artemisinin at therapeutic levels.
They said the lettuce containing the drug could be freeze dried, ground into a powder, and put into capsules for cost-effective delivery.
“Plant and animal science are increasingly coming together,” Dr Kumar said. “In the near future, you will see more drugs produced inside plants will be commercialized to reduce the drug cost.”
Improving cognitive function in cancer survivors
chemotherapy
Photo by Rhoda Baer
A new study suggests a computer program known as InsightTM may help improve cognitive function and overall well-being in cancer survivors.
The study included subjects who reported persistent problems with concentration and/or memory after receiving chemotherapy.
Using the Insight program significantly improved the subjects’ self-reported cognitive function and lowered their levels of anxiety, depression, fatigue, and stress.
However, there was no significant difference in the results of objective neuropsychological function tests between subjects who used the Insight program and subjects who received standard care.
These results were published in the Journal of Clinical Oncology.
“To the best of our knowledge, this is the largest cognitive intervention study that has shown a benefit for patients who are reporting persistent cognitive symptoms following chemotherapy,” said study author Victoria J. Bray, MD, of the University of Sydney in Australia.
About the study
Dr Bray and her colleagues enrolled 242 adult cancer survivors in Australia who had completed at least 3 cycles of chemotherapy in the prior 6 months to 60 months and reported persistent cognitive symptoms.
The subjects’ median age was 53 (range, 23 to 74). Nearly all were women (95%), and 89% had survived breast cancer.
At the beginning of the study, all subjects received a personalized, 30-minute telephone consultation that provided tips and strategies for coping with cognitive problems in daily life.
Subjects were then randomized to Insight (used at home) or standard oncology care per their treating physician.
The primary outcome of the study was self-reported cognitive function, which was assessed using a validated questionnaire known as FACT-COG. It evaluates perceived cognitive impairments, perceived cognitive abilities, and the impact of perceived cognitive impairment on quality of life.
Separate measures were used to evaluate objective neuropsychological function, anxiety/depression, fatigue, and stress.
About the intervention
Dr Bray and her colleagues described Insight as a neurocognitive learning program that uses exercises intended to improve cognition through speed and accuracy of information processing.
The program targets visual precision, divided attention, working memory, field of view, and visual processing speed, which are frequently affected in patients with cancer.
Subjects received the Insight program in disc form and were advised to use the program for 40 minutes 4 times a week for 15 weeks (40 hours total). The program had a built-in measure that could determine subjects’ compliance.
Key findings
Self-reported cognitive function was significantly better in the Insight group than the standard care group, both at the end of the 15-week program and 6 months later.
In addition, Insight participants had significantly lower levels of anxiety, depression, and fatigue immediately after the intervention, significant improvements in quality of life at 6 months, and significant improvements in stress at both time points.
Results of objective neuropsychological function tests were not significantly different between the Insight group and the standard care group, either immediately after the intervention or at 6 months.
Next steps
The researchers said longer follow-up is needed to determine if the effects of the Insight program are long-lasting. And there are still a number of other unanswered questions to be addressed in future research.
For one, it is unclear which method of delivering cognitive rehabilitation is better. A self-directed program such as this one may be suitable for some cancer survivors, while a group-based program may work better for others. It’s also unclear what the ideal duration and “dose” of cognitive training should be.
“If we could identify patients who are at risk of cognitive impairment, we could intervene earlier and possibly achieve even better results,” Dr Bray said. “We would also like to explore whether there is added benefit from combining cognitive training with physical exercise.”

chemotherapy
Photo by Rhoda Baer
A new study suggests a computer program known as InsightTM may help improve cognitive function and overall well-being in cancer survivors.
The study included subjects who reported persistent problems with concentration and/or memory after receiving chemotherapy.
Using the Insight program significantly improved the subjects’ self-reported cognitive function and lowered their levels of anxiety, depression, fatigue, and stress.
However, there was no significant difference in the results of objective neuropsychological function tests between subjects who used the Insight program and subjects who received standard care.
These results were published in the Journal of Clinical Oncology.
“To the best of our knowledge, this is the largest cognitive intervention study that has shown a benefit for patients who are reporting persistent cognitive symptoms following chemotherapy,” said study author Victoria J. Bray, MD, of the University of Sydney in Australia.
About the study
Dr Bray and her colleagues enrolled 242 adult cancer survivors in Australia who had completed at least 3 cycles of chemotherapy in the prior 6 months to 60 months and reported persistent cognitive symptoms.
The subjects’ median age was 53 (range, 23 to 74). Nearly all were women (95%), and 89% had survived breast cancer.
At the beginning of the study, all subjects received a personalized, 30-minute telephone consultation that provided tips and strategies for coping with cognitive problems in daily life.
Subjects were then randomized to Insight (used at home) or standard oncology care per their treating physician.
The primary outcome of the study was self-reported cognitive function, which was assessed using a validated questionnaire known as FACT-COG. It evaluates perceived cognitive impairments, perceived cognitive abilities, and the impact of perceived cognitive impairment on quality of life.
Separate measures were used to evaluate objective neuropsychological function, anxiety/depression, fatigue, and stress.
About the intervention
Dr Bray and her colleagues described Insight as a neurocognitive learning program that uses exercises intended to improve cognition through speed and accuracy of information processing.
The program targets visual precision, divided attention, working memory, field of view, and visual processing speed, which are frequently affected in patients with cancer.
Subjects received the Insight program in disc form and were advised to use the program for 40 minutes 4 times a week for 15 weeks (40 hours total). The program had a built-in measure that could determine subjects’ compliance.
Key findings
Self-reported cognitive function was significantly better in the Insight group than the standard care group, both at the end of the 15-week program and 6 months later.
In addition, Insight participants had significantly lower levels of anxiety, depression, and fatigue immediately after the intervention, significant improvements in quality of life at 6 months, and significant improvements in stress at both time points.
Results of objective neuropsychological function tests were not significantly different between the Insight group and the standard care group, either immediately after the intervention or at 6 months.
Next steps
The researchers said longer follow-up is needed to determine if the effects of the Insight program are long-lasting. And there are still a number of other unanswered questions to be addressed in future research.
For one, it is unclear which method of delivering cognitive rehabilitation is better. A self-directed program such as this one may be suitable for some cancer survivors, while a group-based program may work better for others. It’s also unclear what the ideal duration and “dose” of cognitive training should be.
“If we could identify patients who are at risk of cognitive impairment, we could intervene earlier and possibly achieve even better results,” Dr Bray said. “We would also like to explore whether there is added benefit from combining cognitive training with physical exercise.”

chemotherapy
Photo by Rhoda Baer
A new study suggests a computer program known as InsightTM may help improve cognitive function and overall well-being in cancer survivors.
The study included subjects who reported persistent problems with concentration and/or memory after receiving chemotherapy.
Using the Insight program significantly improved the subjects’ self-reported cognitive function and lowered their levels of anxiety, depression, fatigue, and stress.
However, there was no significant difference in the results of objective neuropsychological function tests between subjects who used the Insight program and subjects who received standard care.
These results were published in the Journal of Clinical Oncology.
“To the best of our knowledge, this is the largest cognitive intervention study that has shown a benefit for patients who are reporting persistent cognitive symptoms following chemotherapy,” said study author Victoria J. Bray, MD, of the University of Sydney in Australia.
About the study
Dr Bray and her colleagues enrolled 242 adult cancer survivors in Australia who had completed at least 3 cycles of chemotherapy in the prior 6 months to 60 months and reported persistent cognitive symptoms.
The subjects’ median age was 53 (range, 23 to 74). Nearly all were women (95%), and 89% had survived breast cancer.
At the beginning of the study, all subjects received a personalized, 30-minute telephone consultation that provided tips and strategies for coping with cognitive problems in daily life.
Subjects were then randomized to Insight (used at home) or standard oncology care per their treating physician.
The primary outcome of the study was self-reported cognitive function, which was assessed using a validated questionnaire known as FACT-COG. It evaluates perceived cognitive impairments, perceived cognitive abilities, and the impact of perceived cognitive impairment on quality of life.
Separate measures were used to evaluate objective neuropsychological function, anxiety/depression, fatigue, and stress.
About the intervention
Dr Bray and her colleagues described Insight as a neurocognitive learning program that uses exercises intended to improve cognition through speed and accuracy of information processing.
The program targets visual precision, divided attention, working memory, field of view, and visual processing speed, which are frequently affected in patients with cancer.
Subjects received the Insight program in disc form and were advised to use the program for 40 minutes 4 times a week for 15 weeks (40 hours total). The program had a built-in measure that could determine subjects’ compliance.
Key findings
Self-reported cognitive function was significantly better in the Insight group than the standard care group, both at the end of the 15-week program and 6 months later.
In addition, Insight participants had significantly lower levels of anxiety, depression, and fatigue immediately after the intervention, significant improvements in quality of life at 6 months, and significant improvements in stress at both time points.
Results of objective neuropsychological function tests were not significantly different between the Insight group and the standard care group, either immediately after the intervention or at 6 months.
Next steps
The researchers said longer follow-up is needed to determine if the effects of the Insight program are long-lasting. And there are still a number of other unanswered questions to be addressed in future research.
For one, it is unclear which method of delivering cognitive rehabilitation is better. A self-directed program such as this one may be suitable for some cancer survivors, while a group-based program may work better for others. It’s also unclear what the ideal duration and “dose” of cognitive training should be.
“If we could identify patients who are at risk of cognitive impairment, we could intervene earlier and possibly achieve even better results,” Dr Bray said. “We would also like to explore whether there is added benefit from combining cognitive training with physical exercise.”
Team maps genomic landscape of CBF-AML

whole-genome sequencing
Photo courtesy of the US
Food and Drug Administration
Whole-genome and whole-exome sequencing has provided new insight into the pathogenesis and development of core-binding factor acute myeloid leukemia (CBF-AML), according to researchers.
The team said their work has revealed “dramatic” differences in the genomic landscape of CBF-AMLs that contribute to the diversity of this disease.
The researchers reported their findings in Nature Genetics.
“We set out to understand the genetic variations that contribute to the development of CBF-AML using whole-exome and whole-genome sequencing,” said study author Jeffery Klco, MD, PhD, of St. Jude Children’s Research Hospital in Memphis, Tennessee.
“Our goal was to define a detailed mutational landscape to understand better the genetic changes that contribute to disease.”
Dr Klco and his colleagues sequenced samples from 87 children and 78 adults with CBF-AML. Eighty-five of the patients had the RUNX1-RUNX1T1 subtype, and 80 had the CBFB-MYH11 subtype.
Development and relapse
The researchers identified several genes with mutations that may contribute to CBF-AML development, including CCND2, DHX15, ASXL2, ZBTB7A, and MGA.
“Many of the mutations we identified interfered with molecular signaling or epigenetic factors,” said study author Jinghui Zhang, PhD, of St. Jude Children’s Research Hospital.
“Some of the mutations, like ASXL2, are epigenetic regulators that modify the local state of chromatin,” Dr Klco noted. “Others, like ZBTB7A, appear to act like tumor suppressors.”
The researchers also compared mutations present at diagnosis and relapse in an attempt to understand how CBF-AML changes over time.
Their results suggested that KMT2C mutations are associated with relapse. Of the 4 patients in this study who had KMT2C mutations, 3 relapsed in less than 12 months, and the fourth had residual disease after a course of remission-induction therapy.
Similarities and differences
The researchers found a similar mutational landscape in adults and children with CBF-AML but differences between patients with the RUNX1-RUNX1T1 and CBFB-MYH11 subtypes.
NRAS was the most frequently mutated gene in CBF-AMLs, but NRAS mutations were more common in CBFB-MYH11 AML than RUNX1-RUNX1T1 AML. The same was true for mutations in NF1 and WT1.
Patients with both subtypes of CBF-AML had mutations in NRAS, KIT, NF1, WT1, FLT3, KRAS, MGA, TTN, CCND2, KDM6A, PHIP, TET2, HCN1, KMT2C, and SETD2.
But only patients with CBFB-MYH11 AML had mutations in PTPN11.
Only patients with RUNX1-RUNX1T1 AML had mutations in ASXL2, ZBTB7A, EZH2, SMC1A, DHX15, RAD21, CBL, DNM2, CSF3R, GIGYF2, SMC3, and ZNF687.
The researchers said their findings suggest a range of mutations may play roles in CBF-AML, but additional research is needed to confirm their precise function in the disease.
Further studies are already underway to fully evaluate the contributions of the different genes as well as the roles of the newly identified genetic alterations in CBF-AML.

whole-genome sequencing
Photo courtesy of the US
Food and Drug Administration
Whole-genome and whole-exome sequencing has provided new insight into the pathogenesis and development of core-binding factor acute myeloid leukemia (CBF-AML), according to researchers.
The team said their work has revealed “dramatic” differences in the genomic landscape of CBF-AMLs that contribute to the diversity of this disease.
The researchers reported their findings in Nature Genetics.
“We set out to understand the genetic variations that contribute to the development of CBF-AML using whole-exome and whole-genome sequencing,” said study author Jeffery Klco, MD, PhD, of St. Jude Children’s Research Hospital in Memphis, Tennessee.
“Our goal was to define a detailed mutational landscape to understand better the genetic changes that contribute to disease.”
Dr Klco and his colleagues sequenced samples from 87 children and 78 adults with CBF-AML. Eighty-five of the patients had the RUNX1-RUNX1T1 subtype, and 80 had the CBFB-MYH11 subtype.
Development and relapse
The researchers identified several genes with mutations that may contribute to CBF-AML development, including CCND2, DHX15, ASXL2, ZBTB7A, and MGA.
“Many of the mutations we identified interfered with molecular signaling or epigenetic factors,” said study author Jinghui Zhang, PhD, of St. Jude Children’s Research Hospital.
“Some of the mutations, like ASXL2, are epigenetic regulators that modify the local state of chromatin,” Dr Klco noted. “Others, like ZBTB7A, appear to act like tumor suppressors.”
The researchers also compared mutations present at diagnosis and relapse in an attempt to understand how CBF-AML changes over time.
Their results suggested that KMT2C mutations are associated with relapse. Of the 4 patients in this study who had KMT2C mutations, 3 relapsed in less than 12 months, and the fourth had residual disease after a course of remission-induction therapy.
Similarities and differences
The researchers found a similar mutational landscape in adults and children with CBF-AML but differences between patients with the RUNX1-RUNX1T1 and CBFB-MYH11 subtypes.
NRAS was the most frequently mutated gene in CBF-AMLs, but NRAS mutations were more common in CBFB-MYH11 AML than RUNX1-RUNX1T1 AML. The same was true for mutations in NF1 and WT1.
Patients with both subtypes of CBF-AML had mutations in NRAS, KIT, NF1, WT1, FLT3, KRAS, MGA, TTN, CCND2, KDM6A, PHIP, TET2, HCN1, KMT2C, and SETD2.
But only patients with CBFB-MYH11 AML had mutations in PTPN11.
Only patients with RUNX1-RUNX1T1 AML had mutations in ASXL2, ZBTB7A, EZH2, SMC1A, DHX15, RAD21, CBL, DNM2, CSF3R, GIGYF2, SMC3, and ZNF687.
The researchers said their findings suggest a range of mutations may play roles in CBF-AML, but additional research is needed to confirm their precise function in the disease.
Further studies are already underway to fully evaluate the contributions of the different genes as well as the roles of the newly identified genetic alterations in CBF-AML.

whole-genome sequencing
Photo courtesy of the US
Food and Drug Administration
Whole-genome and whole-exome sequencing has provided new insight into the pathogenesis and development of core-binding factor acute myeloid leukemia (CBF-AML), according to researchers.
The team said their work has revealed “dramatic” differences in the genomic landscape of CBF-AMLs that contribute to the diversity of this disease.
The researchers reported their findings in Nature Genetics.
“We set out to understand the genetic variations that contribute to the development of CBF-AML using whole-exome and whole-genome sequencing,” said study author Jeffery Klco, MD, PhD, of St. Jude Children’s Research Hospital in Memphis, Tennessee.
“Our goal was to define a detailed mutational landscape to understand better the genetic changes that contribute to disease.”
Dr Klco and his colleagues sequenced samples from 87 children and 78 adults with CBF-AML. Eighty-five of the patients had the RUNX1-RUNX1T1 subtype, and 80 had the CBFB-MYH11 subtype.
Development and relapse
The researchers identified several genes with mutations that may contribute to CBF-AML development, including CCND2, DHX15, ASXL2, ZBTB7A, and MGA.
“Many of the mutations we identified interfered with molecular signaling or epigenetic factors,” said study author Jinghui Zhang, PhD, of St. Jude Children’s Research Hospital.
“Some of the mutations, like ASXL2, are epigenetic regulators that modify the local state of chromatin,” Dr Klco noted. “Others, like ZBTB7A, appear to act like tumor suppressors.”
The researchers also compared mutations present at diagnosis and relapse in an attempt to understand how CBF-AML changes over time.
Their results suggested that KMT2C mutations are associated with relapse. Of the 4 patients in this study who had KMT2C mutations, 3 relapsed in less than 12 months, and the fourth had residual disease after a course of remission-induction therapy.
Similarities and differences
The researchers found a similar mutational landscape in adults and children with CBF-AML but differences between patients with the RUNX1-RUNX1T1 and CBFB-MYH11 subtypes.
NRAS was the most frequently mutated gene in CBF-AMLs, but NRAS mutations were more common in CBFB-MYH11 AML than RUNX1-RUNX1T1 AML. The same was true for mutations in NF1 and WT1.
Patients with both subtypes of CBF-AML had mutations in NRAS, KIT, NF1, WT1, FLT3, KRAS, MGA, TTN, CCND2, KDM6A, PHIP, TET2, HCN1, KMT2C, and SETD2.
But only patients with CBFB-MYH11 AML had mutations in PTPN11.
Only patients with RUNX1-RUNX1T1 AML had mutations in ASXL2, ZBTB7A, EZH2, SMC1A, DHX15, RAD21, CBL, DNM2, CSF3R, GIGYF2, SMC3, and ZNF687.
The researchers said their findings suggest a range of mutations may play roles in CBF-AML, but additional research is needed to confirm their precise function in the disease.
Further studies are already underway to fully evaluate the contributions of the different genes as well as the roles of the newly identified genetic alterations in CBF-AML.