User login
Docs may be uninformed about biosimilars
(filgrastim-sndz), the first
biosimilar approved in the US
© Sandoz Inc. 2015
WASHINGTON, DC—A survey of roughly 1200 US physicians has revealed gaps in their knowledge about biosimilars.
The results showed that some physicians had difficulty defining biosimilars, did not fully understand how the US Food and Drug Administration (FDA) approves biosimilars, and may not know how to prescribe biosimilars appropriately.
The results were published in Advances in Therapy and presented at the DIA Biosimilars 2016 conference.
The 19-question survey was created by the Biosimilars Forum and conducted by SERMO, a global social network organization for physicians, from November 20, 2015, to January 4, 2016.
Responses were obtained from 1201 US physicians across specialties that are high prescribers of biologics, including hematologist-oncologists, dermatologists, gastroenterologists, medical oncologists, nephrologists, and rheumatologists.
Knowledge gaps
The survey revealed 5 major knowledge gaps regarding biosimilars:
- Defining biologics, biosimilars, and biosimilarity1
- Understanding the approval process and the FDA’s use of “totality of evidence” to evaluate biosimilars
- Understanding that the safety profile of a biosimilar is expected to be the same as that of the originator biologic
- Understanding how decisions are made by the FDA for extrapolation2 of indications
- Defining interchangeability3 and the related rules regarding pharmacy-level substitution.
“With 4 biosimilars approved by the FDA and more than 60 in development, the survey highlights the need for greater biosimilars education for physicians and healthcare professionals,” said study author Hillel Cohen, PhD, executive director of scientific affairs at Sandoz Inc.
“Education will help physicians and healthcare professionals have a better understanding and knowledge of biosimilars so that they feel comfortable about administering biosimilars to patients when appropriate.”
FDA approval
Most of the physicians surveyed did not clearly understand the concept of extrapolation as applied to biosimilars. Only 12% of survey respondents said they trust extrapolation of the studied biosimilar indication(s) as the basis to obtain approval of other licensed indications of the originator product.
Almost 60% of respondents correctly understood that, to be approved as “interchangeable” with the originator, a biosimilar must be shown to be safe and effective for back-and-forth switching, with no negative impacts to safety or efficacy.
However, almost 80% of respondents did not agree, or potentially did not realize, that an FDA designation of “interchangeable” may enable a pharmacist to switch between the originator biologic and biosimilar and vice versa.
A little more than half of physicians surveyed knew that, for a biosimilar to be approved, the FDA must find the biosimilar to be equally effective (62.3%) and safe (57.2%) when compared to the originator biologic.
Most physicians surveyed (74.5%) said they trust the FDA’s biosimilar approval decisions.
Biosimilar use
Fewer than half of the survey respondents (44.8%) believed that biosimilars will be safe and appropriate for use in existing patients as well as naïve patients.
Overall, respondents expressed a general agreement that switching an existing patient to a biosimilar may be appropriate, with 9 in 10 (91%) saying they would consider switching a patient from an originator biologic to a biosimilar as an effective alternative to the originator if it would help the patient have better access to his/her medications.
In addition, 82.2% of respondents believed biosimilars will expand treatment options and provide savings to patients and the healthcare system.
“The Biosimilars Forum launched the Partnership for Biosimilar Education and Access to provide evidence-based education for healthcare professionals, patients, and the public,” Dr Cohen said. “The forum will use the survey results to provide education on the key concepts of biosimilars as we advance our mission to encourage awareness, access, and adoption of these important medicines.”
The Biosimilars Forum intends to conduct a similar survey in 2 to 3 years with the same design in order to monitor trends in the awareness, knowledge, and perceptions of biosimilars over time. ![]()
1. A biosimilar medicine is a biologic that is highly similar to an FDA-approved biological medicine, known as an originator biologic. A biosimilar must (1) be highly similar to the originator biologic, notwithstanding minor differences in clinically inactive components, and (2) have no clinically meaningful differences in safety or effectiveness compared to the originator biologic. Biosimilars Forum. (2016, March 28). Frequently Asked Questions. Retrieved from: http://www.biosimilarsforum.org/sites/default/files/uploads/biosimilars_faqs_032816opt_0.pdf
2. Extrapolation is the process by which a biosimilar may be approved for one or more indications for which its reference biological product is licensed but for which there was no head-to-head clinical comparison. This approval is based on the extrapolation of the totality of data obtained with the biosimilar molecule in direct comparison to the originator biologic. Every indication for which extrapolation is sought must be scientifically justified. Biosimilars Forum. (2016, March 28). Frequently Asked Questions. Retrieved from: http://www.biosimilarsforum.org/sites/default/files/uploads/biosimilars_faqs_032816opt_0.pdf
3. An interchangeable biologic is a biosimilar that has been demonstrated to produce the same clinical result as its originator biologic in any given patient. In addition, for a product that is administered multiple times to an individual, the risk in terms of safety or diminished efficacy of alternating between use of the interchangeable biologic and its originator biologic is not greater than the risk of using the originator biologic alone. Designation of interchangeability requires additional supporting evidence, which will be further defined by the FDA. An interchangeable biologic may be substituted by a pharmacist for the originator biologic or vice-versa without the intervention of the healthcare professional who wrote the prescription. State laws govern the substitution process, and communication to the healthcare provider who wrote the prescription may be required after the product is dispensed. Biosimilars Forum. (2016, March 28). Frequently Asked Questions. Retrieved from: http://www.biosimilarsforum.org/sites/default/files/uploads/biosimilars_faqs_032816opt_0.pdf

(filgrastim-sndz), the first
biosimilar approved in the US
© Sandoz Inc. 2015
WASHINGTON, DC—A survey of roughly 1200 US physicians has revealed gaps in their knowledge about biosimilars.
The results showed that some physicians had difficulty defining biosimilars, did not fully understand how the US Food and Drug Administration (FDA) approves biosimilars, and may not know how to prescribe biosimilars appropriately.
The results were published in Advances in Therapy and presented at the DIA Biosimilars 2016 conference.
The 19-question survey was created by the Biosimilars Forum and conducted by SERMO, a global social network organization for physicians, from November 20, 2015, to January 4, 2016.
Responses were obtained from 1201 US physicians across specialties that are high prescribers of biologics, including hematologist-oncologists, dermatologists, gastroenterologists, medical oncologists, nephrologists, and rheumatologists.
Knowledge gaps
The survey revealed 5 major knowledge gaps regarding biosimilars:
- Defining biologics, biosimilars, and biosimilarity1
- Understanding the approval process and the FDA’s use of “totality of evidence” to evaluate biosimilars
- Understanding that the safety profile of a biosimilar is expected to be the same as that of the originator biologic
- Understanding how decisions are made by the FDA for extrapolation2 of indications
- Defining interchangeability3 and the related rules regarding pharmacy-level substitution.
“With 4 biosimilars approved by the FDA and more than 60 in development, the survey highlights the need for greater biosimilars education for physicians and healthcare professionals,” said study author Hillel Cohen, PhD, executive director of scientific affairs at Sandoz Inc.
“Education will help physicians and healthcare professionals have a better understanding and knowledge of biosimilars so that they feel comfortable about administering biosimilars to patients when appropriate.”
FDA approval
Most of the physicians surveyed did not clearly understand the concept of extrapolation as applied to biosimilars. Only 12% of survey respondents said they trust extrapolation of the studied biosimilar indication(s) as the basis to obtain approval of other licensed indications of the originator product.
Almost 60% of respondents correctly understood that, to be approved as “interchangeable” with the originator, a biosimilar must be shown to be safe and effective for back-and-forth switching, with no negative impacts to safety or efficacy.
However, almost 80% of respondents did not agree, or potentially did not realize, that an FDA designation of “interchangeable” may enable a pharmacist to switch between the originator biologic and biosimilar and vice versa.
A little more than half of physicians surveyed knew that, for a biosimilar to be approved, the FDA must find the biosimilar to be equally effective (62.3%) and safe (57.2%) when compared to the originator biologic.
Most physicians surveyed (74.5%) said they trust the FDA’s biosimilar approval decisions.
Biosimilar use
Fewer than half of the survey respondents (44.8%) believed that biosimilars will be safe and appropriate for use in existing patients as well as naïve patients.
Overall, respondents expressed a general agreement that switching an existing patient to a biosimilar may be appropriate, with 9 in 10 (91%) saying they would consider switching a patient from an originator biologic to a biosimilar as an effective alternative to the originator if it would help the patient have better access to his/her medications.
In addition, 82.2% of respondents believed biosimilars will expand treatment options and provide savings to patients and the healthcare system.
“The Biosimilars Forum launched the Partnership for Biosimilar Education and Access to provide evidence-based education for healthcare professionals, patients, and the public,” Dr Cohen said. “The forum will use the survey results to provide education on the key concepts of biosimilars as we advance our mission to encourage awareness, access, and adoption of these important medicines.”
The Biosimilars Forum intends to conduct a similar survey in 2 to 3 years with the same design in order to monitor trends in the awareness, knowledge, and perceptions of biosimilars over time. ![]()
1. A biosimilar medicine is a biologic that is highly similar to an FDA-approved biological medicine, known as an originator biologic. A biosimilar must (1) be highly similar to the originator biologic, notwithstanding minor differences in clinically inactive components, and (2) have no clinically meaningful differences in safety or effectiveness compared to the originator biologic. Biosimilars Forum. (2016, March 28). Frequently Asked Questions. Retrieved from: http://www.biosimilarsforum.org/sites/default/files/uploads/biosimilars_faqs_032816opt_0.pdf
2. Extrapolation is the process by which a biosimilar may be approved for one or more indications for which its reference biological product is licensed but for which there was no head-to-head clinical comparison. This approval is based on the extrapolation of the totality of data obtained with the biosimilar molecule in direct comparison to the originator biologic. Every indication for which extrapolation is sought must be scientifically justified. Biosimilars Forum. (2016, March 28). Frequently Asked Questions. Retrieved from: http://www.biosimilarsforum.org/sites/default/files/uploads/biosimilars_faqs_032816opt_0.pdf
3. An interchangeable biologic is a biosimilar that has been demonstrated to produce the same clinical result as its originator biologic in any given patient. In addition, for a product that is administered multiple times to an individual, the risk in terms of safety or diminished efficacy of alternating between use of the interchangeable biologic and its originator biologic is not greater than the risk of using the originator biologic alone. Designation of interchangeability requires additional supporting evidence, which will be further defined by the FDA. An interchangeable biologic may be substituted by a pharmacist for the originator biologic or vice-versa without the intervention of the healthcare professional who wrote the prescription. State laws govern the substitution process, and communication to the healthcare provider who wrote the prescription may be required after the product is dispensed. Biosimilars Forum. (2016, March 28). Frequently Asked Questions. Retrieved from: http://www.biosimilarsforum.org/sites/default/files/uploads/biosimilars_faqs_032816opt_0.pdf

(filgrastim-sndz), the first
biosimilar approved in the US
© Sandoz Inc. 2015
WASHINGTON, DC—A survey of roughly 1200 US physicians has revealed gaps in their knowledge about biosimilars.
The results showed that some physicians had difficulty defining biosimilars, did not fully understand how the US Food and Drug Administration (FDA) approves biosimilars, and may not know how to prescribe biosimilars appropriately.
The results were published in Advances in Therapy and presented at the DIA Biosimilars 2016 conference.
The 19-question survey was created by the Biosimilars Forum and conducted by SERMO, a global social network organization for physicians, from November 20, 2015, to January 4, 2016.
Responses were obtained from 1201 US physicians across specialties that are high prescribers of biologics, including hematologist-oncologists, dermatologists, gastroenterologists, medical oncologists, nephrologists, and rheumatologists.
Knowledge gaps
The survey revealed 5 major knowledge gaps regarding biosimilars:
- Defining biologics, biosimilars, and biosimilarity1
- Understanding the approval process and the FDA’s use of “totality of evidence” to evaluate biosimilars
- Understanding that the safety profile of a biosimilar is expected to be the same as that of the originator biologic
- Understanding how decisions are made by the FDA for extrapolation2 of indications
- Defining interchangeability3 and the related rules regarding pharmacy-level substitution.
“With 4 biosimilars approved by the FDA and more than 60 in development, the survey highlights the need for greater biosimilars education for physicians and healthcare professionals,” said study author Hillel Cohen, PhD, executive director of scientific affairs at Sandoz Inc.
“Education will help physicians and healthcare professionals have a better understanding and knowledge of biosimilars so that they feel comfortable about administering biosimilars to patients when appropriate.”
FDA approval
Most of the physicians surveyed did not clearly understand the concept of extrapolation as applied to biosimilars. Only 12% of survey respondents said they trust extrapolation of the studied biosimilar indication(s) as the basis to obtain approval of other licensed indications of the originator product.
Almost 60% of respondents correctly understood that, to be approved as “interchangeable” with the originator, a biosimilar must be shown to be safe and effective for back-and-forth switching, with no negative impacts to safety or efficacy.
However, almost 80% of respondents did not agree, or potentially did not realize, that an FDA designation of “interchangeable” may enable a pharmacist to switch between the originator biologic and biosimilar and vice versa.
A little more than half of physicians surveyed knew that, for a biosimilar to be approved, the FDA must find the biosimilar to be equally effective (62.3%) and safe (57.2%) when compared to the originator biologic.
Most physicians surveyed (74.5%) said they trust the FDA’s biosimilar approval decisions.
Biosimilar use
Fewer than half of the survey respondents (44.8%) believed that biosimilars will be safe and appropriate for use in existing patients as well as naïve patients.
Overall, respondents expressed a general agreement that switching an existing patient to a biosimilar may be appropriate, with 9 in 10 (91%) saying they would consider switching a patient from an originator biologic to a biosimilar as an effective alternative to the originator if it would help the patient have better access to his/her medications.
In addition, 82.2% of respondents believed biosimilars will expand treatment options and provide savings to patients and the healthcare system.
“The Biosimilars Forum launched the Partnership for Biosimilar Education and Access to provide evidence-based education for healthcare professionals, patients, and the public,” Dr Cohen said. “The forum will use the survey results to provide education on the key concepts of biosimilars as we advance our mission to encourage awareness, access, and adoption of these important medicines.”
The Biosimilars Forum intends to conduct a similar survey in 2 to 3 years with the same design in order to monitor trends in the awareness, knowledge, and perceptions of biosimilars over time. ![]()
1. A biosimilar medicine is a biologic that is highly similar to an FDA-approved biological medicine, known as an originator biologic. A biosimilar must (1) be highly similar to the originator biologic, notwithstanding minor differences in clinically inactive components, and (2) have no clinically meaningful differences in safety or effectiveness compared to the originator biologic. Biosimilars Forum. (2016, March 28). Frequently Asked Questions. Retrieved from: http://www.biosimilarsforum.org/sites/default/files/uploads/biosimilars_faqs_032816opt_0.pdf
2. Extrapolation is the process by which a biosimilar may be approved for one or more indications for which its reference biological product is licensed but for which there was no head-to-head clinical comparison. This approval is based on the extrapolation of the totality of data obtained with the biosimilar molecule in direct comparison to the originator biologic. Every indication for which extrapolation is sought must be scientifically justified. Biosimilars Forum. (2016, March 28). Frequently Asked Questions. Retrieved from: http://www.biosimilarsforum.org/sites/default/files/uploads/biosimilars_faqs_032816opt_0.pdf
3. An interchangeable biologic is a biosimilar that has been demonstrated to produce the same clinical result as its originator biologic in any given patient. In addition, for a product that is administered multiple times to an individual, the risk in terms of safety or diminished efficacy of alternating between use of the interchangeable biologic and its originator biologic is not greater than the risk of using the originator biologic alone. Designation of interchangeability requires additional supporting evidence, which will be further defined by the FDA. An interchangeable biologic may be substituted by a pharmacist for the originator biologic or vice-versa without the intervention of the healthcare professional who wrote the prescription. State laws govern the substitution process, and communication to the healthcare provider who wrote the prescription may be required after the product is dispensed. Biosimilars Forum. (2016, March 28). Frequently Asked Questions. Retrieved from: http://www.biosimilarsforum.org/sites/default/files/uploads/biosimilars_faqs_032816opt_0.pdf
Study provides new insight into RBC production

Andes who suffers from
chronic mountain sickness
Photo courtesy of
UC San Diego Health
Findings from a study of people living at high altitude have implications for the treatment of red blood cell (RBC) disorders such as anemia and polycythemia, according to researchers.
To better understand why some people adapt well to life at high altitude while others don’t, the researchers studied RBCs derived from representatives of both groups who were living in the Andes Mountains.
The study revealed that high-altitude dwellers prone to chronic mountain sickness produce massive amounts of RBCs thanks to overproduction of the enzyme SENP1.
The researchers reported these findings in the Journal of Experimental Medicine.
“In addition to improving the health of millions of people around the world who live above 8000 feet, information on how Andeans have adapted—or not adapted—to high-altitude life might teach us how to speed up red blood cell production at lower altitudes, such as in anemia or when blood transfusions are needed rapidly,” said study author Gabriel Haddad, MD, of the University of California San Diego School of Medicine.
Dr Haddad and his colleagues noted that chronic mountain sickness affects approximately 20% of people who live at high altitudes, and a critical aspect of the condition is polycythemia.
Some extra RBCs can be beneficial in high-altitude, low-oxygen environments by helping to keep blood oxygenated. However, too many RBCs can increase the risk of heart attack and stroke, even in young adults.
For this study, the researchers collected skin cells from people living in the Andes Mountains—4 who were healthy and 5 who suffer from chronic mountain sickness—plus an additional 3 healthy people who live at sea level and served as controls.
To produce enough RBCs from each participant to study them in the lab, the researchers converted the skin cells into induced pluripotent stem cells (iPSCs).
Then, adding a cocktail of growth factors and other molecules, the team coaxed the iPSCs to differentiate into RBCs. Multiple samples were tested for each person, for a total of at least 24 iPSC lines.
The researchers exposed the RBCs to low-oxygen conditions that mimic high altitude—5% oxygen—for 3 weeks.
RBCs from healthy sea-level or high-altitude-dwelling donors increased a little or not at all. In contrast, RBC counts from high-altitude dwellers with chronic mountain sickness increased 60-fold.
This result led the researchers to question why people with chronic mountain sickness produce so many extra RBCs in response to low oxygen.
In a previous study, the team had compared the genomes of high-altitude dwellers with and without chronic mountain sickness. This revealed a gene that varied between the 2 groups—SENP1, which is increased in low-oxygen situations in people with chronic mountain sickness but not in healthy individuals.
In the current study, the researchers set out to determine if SENP1 plays a role in high-altitude adaptation.
The team inhibited the SENP1 gene in iPSCs from patients with chronic mountain sickness. As a result, excessive RBC production was reduced by more than 90%.
When the researchers added extra SENP1 to healthy, adapted highlander iPSCs, RBC production increased 30-fold, nearly recapitulating that seen in patients with chronic mountains sickness.
Further experiments revealed how SENP1 affects RBC production. Elevated levels of the enzyme in chronic mountain sickness boost levels of several other proteins that promote cell division and survival, including VEGF, GATA1, and Bcl-xL.
“We’re interested in determining the early steps in this process—how low oxygen triggers SENP1 in the first place,” said study author Priti Azad, PhD, of the University of California San Diego School of Medicine.
“We are also investigating how existing altitude sickness medications, such as Diamox, work and whether or not it’s through this same mechanism.” ![]()

Andes who suffers from
chronic mountain sickness
Photo courtesy of
UC San Diego Health
Findings from a study of people living at high altitude have implications for the treatment of red blood cell (RBC) disorders such as anemia and polycythemia, according to researchers.
To better understand why some people adapt well to life at high altitude while others don’t, the researchers studied RBCs derived from representatives of both groups who were living in the Andes Mountains.
The study revealed that high-altitude dwellers prone to chronic mountain sickness produce massive amounts of RBCs thanks to overproduction of the enzyme SENP1.
The researchers reported these findings in the Journal of Experimental Medicine.
“In addition to improving the health of millions of people around the world who live above 8000 feet, information on how Andeans have adapted—or not adapted—to high-altitude life might teach us how to speed up red blood cell production at lower altitudes, such as in anemia or when blood transfusions are needed rapidly,” said study author Gabriel Haddad, MD, of the University of California San Diego School of Medicine.
Dr Haddad and his colleagues noted that chronic mountain sickness affects approximately 20% of people who live at high altitudes, and a critical aspect of the condition is polycythemia.
Some extra RBCs can be beneficial in high-altitude, low-oxygen environments by helping to keep blood oxygenated. However, too many RBCs can increase the risk of heart attack and stroke, even in young adults.
For this study, the researchers collected skin cells from people living in the Andes Mountains—4 who were healthy and 5 who suffer from chronic mountain sickness—plus an additional 3 healthy people who live at sea level and served as controls.
To produce enough RBCs from each participant to study them in the lab, the researchers converted the skin cells into induced pluripotent stem cells (iPSCs).
Then, adding a cocktail of growth factors and other molecules, the team coaxed the iPSCs to differentiate into RBCs. Multiple samples were tested for each person, for a total of at least 24 iPSC lines.
The researchers exposed the RBCs to low-oxygen conditions that mimic high altitude—5% oxygen—for 3 weeks.
RBCs from healthy sea-level or high-altitude-dwelling donors increased a little or not at all. In contrast, RBC counts from high-altitude dwellers with chronic mountain sickness increased 60-fold.
This result led the researchers to question why people with chronic mountain sickness produce so many extra RBCs in response to low oxygen.
In a previous study, the team had compared the genomes of high-altitude dwellers with and without chronic mountain sickness. This revealed a gene that varied between the 2 groups—SENP1, which is increased in low-oxygen situations in people with chronic mountain sickness but not in healthy individuals.
In the current study, the researchers set out to determine if SENP1 plays a role in high-altitude adaptation.
The team inhibited the SENP1 gene in iPSCs from patients with chronic mountain sickness. As a result, excessive RBC production was reduced by more than 90%.
When the researchers added extra SENP1 to healthy, adapted highlander iPSCs, RBC production increased 30-fold, nearly recapitulating that seen in patients with chronic mountains sickness.
Further experiments revealed how SENP1 affects RBC production. Elevated levels of the enzyme in chronic mountain sickness boost levels of several other proteins that promote cell division and survival, including VEGF, GATA1, and Bcl-xL.
“We’re interested in determining the early steps in this process—how low oxygen triggers SENP1 in the first place,” said study author Priti Azad, PhD, of the University of California San Diego School of Medicine.
“We are also investigating how existing altitude sickness medications, such as Diamox, work and whether or not it’s through this same mechanism.” ![]()

Andes who suffers from
chronic mountain sickness
Photo courtesy of
UC San Diego Health
Findings from a study of people living at high altitude have implications for the treatment of red blood cell (RBC) disorders such as anemia and polycythemia, according to researchers.
To better understand why some people adapt well to life at high altitude while others don’t, the researchers studied RBCs derived from representatives of both groups who were living in the Andes Mountains.
The study revealed that high-altitude dwellers prone to chronic mountain sickness produce massive amounts of RBCs thanks to overproduction of the enzyme SENP1.
The researchers reported these findings in the Journal of Experimental Medicine.
“In addition to improving the health of millions of people around the world who live above 8000 feet, information on how Andeans have adapted—or not adapted—to high-altitude life might teach us how to speed up red blood cell production at lower altitudes, such as in anemia or when blood transfusions are needed rapidly,” said study author Gabriel Haddad, MD, of the University of California San Diego School of Medicine.
Dr Haddad and his colleagues noted that chronic mountain sickness affects approximately 20% of people who live at high altitudes, and a critical aspect of the condition is polycythemia.
Some extra RBCs can be beneficial in high-altitude, low-oxygen environments by helping to keep blood oxygenated. However, too many RBCs can increase the risk of heart attack and stroke, even in young adults.
For this study, the researchers collected skin cells from people living in the Andes Mountains—4 who were healthy and 5 who suffer from chronic mountain sickness—plus an additional 3 healthy people who live at sea level and served as controls.
To produce enough RBCs from each participant to study them in the lab, the researchers converted the skin cells into induced pluripotent stem cells (iPSCs).
Then, adding a cocktail of growth factors and other molecules, the team coaxed the iPSCs to differentiate into RBCs. Multiple samples were tested for each person, for a total of at least 24 iPSC lines.
The researchers exposed the RBCs to low-oxygen conditions that mimic high altitude—5% oxygen—for 3 weeks.
RBCs from healthy sea-level or high-altitude-dwelling donors increased a little or not at all. In contrast, RBC counts from high-altitude dwellers with chronic mountain sickness increased 60-fold.
This result led the researchers to question why people with chronic mountain sickness produce so many extra RBCs in response to low oxygen.
In a previous study, the team had compared the genomes of high-altitude dwellers with and without chronic mountain sickness. This revealed a gene that varied between the 2 groups—SENP1, which is increased in low-oxygen situations in people with chronic mountain sickness but not in healthy individuals.
In the current study, the researchers set out to determine if SENP1 plays a role in high-altitude adaptation.
The team inhibited the SENP1 gene in iPSCs from patients with chronic mountain sickness. As a result, excessive RBC production was reduced by more than 90%.
When the researchers added extra SENP1 to healthy, adapted highlander iPSCs, RBC production increased 30-fold, nearly recapitulating that seen in patients with chronic mountains sickness.
Further experiments revealed how SENP1 affects RBC production. Elevated levels of the enzyme in chronic mountain sickness boost levels of several other proteins that promote cell division and survival, including VEGF, GATA1, and Bcl-xL.
“We’re interested in determining the early steps in this process—how low oxygen triggers SENP1 in the first place,” said study author Priti Azad, PhD, of the University of California San Diego School of Medicine.
“We are also investigating how existing altitude sickness medications, such as Diamox, work and whether or not it’s through this same mechanism.” ![]()
Antiviral appears active against tough-to-treat CMV
NEW ORLEANS—The antiviral agent maribavir has shown activity against cytomegalovirus (CMV) in a phase 2 trial.
The trial included transplant recipients with a CMV infection that was resistant or refractory to prior treatment with (val)ganciclovir or foscarnet.
Sixty-seven percent of patients treated with varying doses of maribavir for up to 24 weeks had no detectable levels of CMV in their plasma within 6 weeks of starting treatment.
However, CMV infection did recur in some of these patients.
Dysgeusia was the most commonly reported treatment-emergent adverse event (AE), and one patient died of an AE that was considered possibly related to maribavir.
These results were presented at IDWeek 2016 (abstract 78). The study was sponsored by Shire.
“Cytomegalovirus infection that is resistant or refractory to standard therapy in transplant patients is associated with significant complications and high mortality,” said study investigator Genovefa Papanicolaou, MD, of Memorial Sloan Kettering Cancer Center in New York, New York.
“The phase 2 findings support further research to confirm these results among patients who have limited options to combat the infection.”
Maribavir is a member of a class of drugs called benzimidazole ribosides. Maribavir is thought to inhibit viral DNA assembly and egress of viral capsids from the nucleus of infected cells. The drug has not been shown to affect the maturation of viral DNA or affect the viral DNA polymerase.
This phase 2 study of maribavir included 120 patients ages 12 and older with CMV infection (≥1000 DNA copies/mL of blood plasma) that was resistant or refractory to (val)ganciclovir or foscarnet.
Forty-seven of the patients had received a hematopoietic stem cell transplant, and 73 had a solid organ transplant. The patients’ median age was 55 (range, 18 to 74).
The patients were randomized to 1 of 3 twice-daily oral doses of maribavir—400 mg, 800 mg, or 1200 mg—for up to 24 weeks of treatment.
Efficacy
The study’s primary efficacy endpoint was the proportion of patients with confirmed undetectable plasma CMV DNA within 6 weeks of treatment.
Overall, 67% (80/120) of patients met the primary efficacy endpoint. This included 70% (n=28) of patients in the 400 mg group, 63% (n=25) in the 800 mg group, and 67% (n=27) in the 1200 mg group.
CMV infection recurred in 30 patients, including 7 in the 400 mg group, 11 in the 800 mg group, and 12 in the 1200 mg group.
Safety
The primary safety analysis was focused on the incidence of treatment-emergent AEs. The incidence was 78% (n=93) overall, including 78% (n=31) in the 400 mg group, 80% (n=32) in the 800 mg group, and 75% (n=30) in the 1200 mg group.
Twenty-seven percent of patients died due to any AE, 1 of which (multi-organ failure) was considered possibly related to maribavir.
Forty-one patients (34%) discontinued treatment with maribavir due to an AE, including 17 patients who discontinued due to CMV infection.
Dysgeusia was the most common treatment-emergent AE and led to treatment discontinuation in 1 patient. Dysgeusia occurred in 65% (n=78) of all patients, including 60% (n=24) in the 400 mg group, 63% (n=25) in the 800 mg group, and 73% (n=29) in the 1200 mg group.
Other treatment-emergent AEs (occurring in at least 20% of patients) for all doses included nausea, vomiting, CMV infection, diarrhea, fatigue, and anemia. Immunosuppressant drug level increases were reported as an AE in 10% of all patients. ![]()

NEW ORLEANS—The antiviral agent maribavir has shown activity against cytomegalovirus (CMV) in a phase 2 trial.
The trial included transplant recipients with a CMV infection that was resistant or refractory to prior treatment with (val)ganciclovir or foscarnet.
Sixty-seven percent of patients treated with varying doses of maribavir for up to 24 weeks had no detectable levels of CMV in their plasma within 6 weeks of starting treatment.
However, CMV infection did recur in some of these patients.
Dysgeusia was the most commonly reported treatment-emergent adverse event (AE), and one patient died of an AE that was considered possibly related to maribavir.
These results were presented at IDWeek 2016 (abstract 78). The study was sponsored by Shire.
“Cytomegalovirus infection that is resistant or refractory to standard therapy in transplant patients is associated with significant complications and high mortality,” said study investigator Genovefa Papanicolaou, MD, of Memorial Sloan Kettering Cancer Center in New York, New York.
“The phase 2 findings support further research to confirm these results among patients who have limited options to combat the infection.”
Maribavir is a member of a class of drugs called benzimidazole ribosides. Maribavir is thought to inhibit viral DNA assembly and egress of viral capsids from the nucleus of infected cells. The drug has not been shown to affect the maturation of viral DNA or affect the viral DNA polymerase.
This phase 2 study of maribavir included 120 patients ages 12 and older with CMV infection (≥1000 DNA copies/mL of blood plasma) that was resistant or refractory to (val)ganciclovir or foscarnet.
Forty-seven of the patients had received a hematopoietic stem cell transplant, and 73 had a solid organ transplant. The patients’ median age was 55 (range, 18 to 74).
The patients were randomized to 1 of 3 twice-daily oral doses of maribavir—400 mg, 800 mg, or 1200 mg—for up to 24 weeks of treatment.
Efficacy
The study’s primary efficacy endpoint was the proportion of patients with confirmed undetectable plasma CMV DNA within 6 weeks of treatment.
Overall, 67% (80/120) of patients met the primary efficacy endpoint. This included 70% (n=28) of patients in the 400 mg group, 63% (n=25) in the 800 mg group, and 67% (n=27) in the 1200 mg group.
CMV infection recurred in 30 patients, including 7 in the 400 mg group, 11 in the 800 mg group, and 12 in the 1200 mg group.
Safety
The primary safety analysis was focused on the incidence of treatment-emergent AEs. The incidence was 78% (n=93) overall, including 78% (n=31) in the 400 mg group, 80% (n=32) in the 800 mg group, and 75% (n=30) in the 1200 mg group.
Twenty-seven percent of patients died due to any AE, 1 of which (multi-organ failure) was considered possibly related to maribavir.
Forty-one patients (34%) discontinued treatment with maribavir due to an AE, including 17 patients who discontinued due to CMV infection.
Dysgeusia was the most common treatment-emergent AE and led to treatment discontinuation in 1 patient. Dysgeusia occurred in 65% (n=78) of all patients, including 60% (n=24) in the 400 mg group, 63% (n=25) in the 800 mg group, and 73% (n=29) in the 1200 mg group.
Other treatment-emergent AEs (occurring in at least 20% of patients) for all doses included nausea, vomiting, CMV infection, diarrhea, fatigue, and anemia. Immunosuppressant drug level increases were reported as an AE in 10% of all patients. ![]()

NEW ORLEANS—The antiviral agent maribavir has shown activity against cytomegalovirus (CMV) in a phase 2 trial.
The trial included transplant recipients with a CMV infection that was resistant or refractory to prior treatment with (val)ganciclovir or foscarnet.
Sixty-seven percent of patients treated with varying doses of maribavir for up to 24 weeks had no detectable levels of CMV in their plasma within 6 weeks of starting treatment.
However, CMV infection did recur in some of these patients.
Dysgeusia was the most commonly reported treatment-emergent adverse event (AE), and one patient died of an AE that was considered possibly related to maribavir.
These results were presented at IDWeek 2016 (abstract 78). The study was sponsored by Shire.
“Cytomegalovirus infection that is resistant or refractory to standard therapy in transplant patients is associated with significant complications and high mortality,” said study investigator Genovefa Papanicolaou, MD, of Memorial Sloan Kettering Cancer Center in New York, New York.
“The phase 2 findings support further research to confirm these results among patients who have limited options to combat the infection.”
Maribavir is a member of a class of drugs called benzimidazole ribosides. Maribavir is thought to inhibit viral DNA assembly and egress of viral capsids from the nucleus of infected cells. The drug has not been shown to affect the maturation of viral DNA or affect the viral DNA polymerase.
This phase 2 study of maribavir included 120 patients ages 12 and older with CMV infection (≥1000 DNA copies/mL of blood plasma) that was resistant or refractory to (val)ganciclovir or foscarnet.
Forty-seven of the patients had received a hematopoietic stem cell transplant, and 73 had a solid organ transplant. The patients’ median age was 55 (range, 18 to 74).
The patients were randomized to 1 of 3 twice-daily oral doses of maribavir—400 mg, 800 mg, or 1200 mg—for up to 24 weeks of treatment.
Efficacy
The study’s primary efficacy endpoint was the proportion of patients with confirmed undetectable plasma CMV DNA within 6 weeks of treatment.
Overall, 67% (80/120) of patients met the primary efficacy endpoint. This included 70% (n=28) of patients in the 400 mg group, 63% (n=25) in the 800 mg group, and 67% (n=27) in the 1200 mg group.
CMV infection recurred in 30 patients, including 7 in the 400 mg group, 11 in the 800 mg group, and 12 in the 1200 mg group.
Safety
The primary safety analysis was focused on the incidence of treatment-emergent AEs. The incidence was 78% (n=93) overall, including 78% (n=31) in the 400 mg group, 80% (n=32) in the 800 mg group, and 75% (n=30) in the 1200 mg group.
Twenty-seven percent of patients died due to any AE, 1 of which (multi-organ failure) was considered possibly related to maribavir.
Forty-one patients (34%) discontinued treatment with maribavir due to an AE, including 17 patients who discontinued due to CMV infection.
Dysgeusia was the most common treatment-emergent AE and led to treatment discontinuation in 1 patient. Dysgeusia occurred in 65% (n=78) of all patients, including 60% (n=24) in the 400 mg group, 63% (n=25) in the 800 mg group, and 73% (n=29) in the 1200 mg group.
Other treatment-emergent AEs (occurring in at least 20% of patients) for all doses included nausea, vomiting, CMV infection, diarrhea, fatigue, and anemia. Immunosuppressant drug level increases were reported as an AE in 10% of all patients. ![]()
Drug elicits responses in heavily pretreated cHL
Photo from Business Wire
COLOGNE—Results from the CheckMate -205 study suggest nivolumab can produce a high objective response rate (ORR) in patients with heavily pretreated classical Hodgkin lymphoma (cHL).
Investigators recently presented results from one of the cohorts in this phase 2 trial—cohort C—which included cHL patients who received nivolumab after undergoing autologous hematopoietic stem cell transplant (auto-HSCT) and receiving treatment with brentuximab vedotin (BV).
At a median follow-up of 8.8 months, the ORR, as assessed by an independent radiologic review committee (IRRC), was 73%.
The median progression-free survival (PFS) was 11.2 months, and the 6-month overall survival (OS) was 94%.
Investigators said the safety profile of nivolumab in this patient population was consistent with previously reported data in patients with cHL, and no new clinically meaningful safety signals were identified.
“These data from cohort C build on existing evidence supporting the benefit of Opdivo [nivolumab] in classical Hodgkin lymphoma patients who have relapsed or progressed after autologous hematopoietic stem cell transplantation and post-transplantation brentuximab vedotin,” said investigator Andreas Engert, MD, of the University Hospital of Cologne in Germany.
“Results from cohort C indicated a benefit with Opdivo regardless of the order of prior treatment with autologous hematopoietic stem cell transplantation and brentuximab vedotin, providing important insights as we continue researching the potential role Opdivo could provide for heavily pretreated classical Hodgkin lymphoma patients.”
The results were presented at the 10th International Symposium on Hodgkin Lymphoma (abstract T022). Abstracts from this meeting have been published in haematologica.
The CheckMate -205 trial is sponsored by Bristol-Myers Squibb.
About the trial
CheckMate -205 is a multi-cohort study in which investigators are evaluating the safety and efficacy of nivolumab in adults with cHL.
Cohort A included cHL patients who had received auto-HSCT and were BV-naïve (n=63). Cohort B included cHL patients who had received auto-HSCT followed by BV (n=80).
Cohort C included cHL patients who had received BV before and/or after auto-HSCT (n=100).
The study also includes a cohort D, which is currently enrolling and evaluating nivolumab in combination with chemotherapy in newly diagnosed, advanced-stage cHL patients who are treatment-naïve (n=50).
Patients enrolled in this trial have received nivolumab at 3 mg/kg intravenously every 2 weeks until disease progression or unacceptable toxicity. In cohort C, patients were also treated until investigator-assessed complete response (CR) lasting 1 year.
The study’s primary endpoint was ORR by IRRC assessment. Secondary and other exploratory endpoints included duration of response by IRRC assessment for CR rate and partial response rate, PFS by IRRC assessment, OS, and safety.
Response
The investigators presented results from cohort C (n=100), which included patients with cHL who had received BV before and/or after auto-HSCT.
At a median follow-up of 8.8 months, ORR per the IRRC was 73% (n=73) overall. The investigators said ORR was consistent across patient subgroups, regardless of the timing of prior BV relative to auto-HSCT.
The ORR was 70% (n=23) in patients who received BV only before auto-HSCT, 72% (n=41) in patients who received BV only after auto-HSCT, and 88% (n=7) in patients who received BV before and after auto-HSCT.
The CR rate was 17% (n=17) overall, 18.2% (n=6) in patients who received BV only before auto-HSCT, 12.3% (n=7) in patients who received BV only after auto-HSCT, and 38% (n=3) in patients who received BV before and after auto-HSCT.
Survival
The median PFS was 11.2 months (range, 8.5 months to not achieved) overall, 11.2 months (range, 8.5 months to not achieved) in patients who received BV only before auto-HSCT, 8.9 months (range, 8.3 months to not achieved) in patients who received BV only after auto-HSCT, and not achieved (range, 5.6 months to not achieved) in patients who received BV before and after auto-HSCT.
The 6-month OS was 93.9% overall, 97% in patients who received BV only before auto-HSCT, 91% in patients who received BV only after auto-HSCT, and 100% in patients who received BV before and after auto-HSCT.
Safety
Treatment-related adverse events (AEs) occurred in 68% of patients between the first dose and 30 days after the last dose of nivolumab. The most common treatment-related AEs were diarrhea, infusion-related reaction, and fatigue (11% each).
Grade 3/4 AEs occurred in 19% of patients. Serious treatment-related AEs were reported in 17% of patients, and treatment-related AEs leading to discontinuation occurred in 6% of patients.
At present, no treatment-related deaths have been reported. ![]()

Photo from Business Wire
COLOGNE—Results from the CheckMate -205 study suggest nivolumab can produce a high objective response rate (ORR) in patients with heavily pretreated classical Hodgkin lymphoma (cHL).
Investigators recently presented results from one of the cohorts in this phase 2 trial—cohort C—which included cHL patients who received nivolumab after undergoing autologous hematopoietic stem cell transplant (auto-HSCT) and receiving treatment with brentuximab vedotin (BV).
At a median follow-up of 8.8 months, the ORR, as assessed by an independent radiologic review committee (IRRC), was 73%.
The median progression-free survival (PFS) was 11.2 months, and the 6-month overall survival (OS) was 94%.
Investigators said the safety profile of nivolumab in this patient population was consistent with previously reported data in patients with cHL, and no new clinically meaningful safety signals were identified.
“These data from cohort C build on existing evidence supporting the benefit of Opdivo [nivolumab] in classical Hodgkin lymphoma patients who have relapsed or progressed after autologous hematopoietic stem cell transplantation and post-transplantation brentuximab vedotin,” said investigator Andreas Engert, MD, of the University Hospital of Cologne in Germany.
“Results from cohort C indicated a benefit with Opdivo regardless of the order of prior treatment with autologous hematopoietic stem cell transplantation and brentuximab vedotin, providing important insights as we continue researching the potential role Opdivo could provide for heavily pretreated classical Hodgkin lymphoma patients.”
The results were presented at the 10th International Symposium on Hodgkin Lymphoma (abstract T022). Abstracts from this meeting have been published in haematologica.
The CheckMate -205 trial is sponsored by Bristol-Myers Squibb.
About the trial
CheckMate -205 is a multi-cohort study in which investigators are evaluating the safety and efficacy of nivolumab in adults with cHL.
Cohort A included cHL patients who had received auto-HSCT and were BV-naïve (n=63). Cohort B included cHL patients who had received auto-HSCT followed by BV (n=80).
Cohort C included cHL patients who had received BV before and/or after auto-HSCT (n=100).
The study also includes a cohort D, which is currently enrolling and evaluating nivolumab in combination with chemotherapy in newly diagnosed, advanced-stage cHL patients who are treatment-naïve (n=50).
Patients enrolled in this trial have received nivolumab at 3 mg/kg intravenously every 2 weeks until disease progression or unacceptable toxicity. In cohort C, patients were also treated until investigator-assessed complete response (CR) lasting 1 year.
The study’s primary endpoint was ORR by IRRC assessment. Secondary and other exploratory endpoints included duration of response by IRRC assessment for CR rate and partial response rate, PFS by IRRC assessment, OS, and safety.
Response
The investigators presented results from cohort C (n=100), which included patients with cHL who had received BV before and/or after auto-HSCT.
At a median follow-up of 8.8 months, ORR per the IRRC was 73% (n=73) overall. The investigators said ORR was consistent across patient subgroups, regardless of the timing of prior BV relative to auto-HSCT.
The ORR was 70% (n=23) in patients who received BV only before auto-HSCT, 72% (n=41) in patients who received BV only after auto-HSCT, and 88% (n=7) in patients who received BV before and after auto-HSCT.
The CR rate was 17% (n=17) overall, 18.2% (n=6) in patients who received BV only before auto-HSCT, 12.3% (n=7) in patients who received BV only after auto-HSCT, and 38% (n=3) in patients who received BV before and after auto-HSCT.
Survival
The median PFS was 11.2 months (range, 8.5 months to not achieved) overall, 11.2 months (range, 8.5 months to not achieved) in patients who received BV only before auto-HSCT, 8.9 months (range, 8.3 months to not achieved) in patients who received BV only after auto-HSCT, and not achieved (range, 5.6 months to not achieved) in patients who received BV before and after auto-HSCT.
The 6-month OS was 93.9% overall, 97% in patients who received BV only before auto-HSCT, 91% in patients who received BV only after auto-HSCT, and 100% in patients who received BV before and after auto-HSCT.
Safety
Treatment-related adverse events (AEs) occurred in 68% of patients between the first dose and 30 days after the last dose of nivolumab. The most common treatment-related AEs were diarrhea, infusion-related reaction, and fatigue (11% each).
Grade 3/4 AEs occurred in 19% of patients. Serious treatment-related AEs were reported in 17% of patients, and treatment-related AEs leading to discontinuation occurred in 6% of patients.
At present, no treatment-related deaths have been reported. ![]()

Photo from Business Wire
COLOGNE—Results from the CheckMate -205 study suggest nivolumab can produce a high objective response rate (ORR) in patients with heavily pretreated classical Hodgkin lymphoma (cHL).
Investigators recently presented results from one of the cohorts in this phase 2 trial—cohort C—which included cHL patients who received nivolumab after undergoing autologous hematopoietic stem cell transplant (auto-HSCT) and receiving treatment with brentuximab vedotin (BV).
At a median follow-up of 8.8 months, the ORR, as assessed by an independent radiologic review committee (IRRC), was 73%.
The median progression-free survival (PFS) was 11.2 months, and the 6-month overall survival (OS) was 94%.
Investigators said the safety profile of nivolumab in this patient population was consistent with previously reported data in patients with cHL, and no new clinically meaningful safety signals were identified.
“These data from cohort C build on existing evidence supporting the benefit of Opdivo [nivolumab] in classical Hodgkin lymphoma patients who have relapsed or progressed after autologous hematopoietic stem cell transplantation and post-transplantation brentuximab vedotin,” said investigator Andreas Engert, MD, of the University Hospital of Cologne in Germany.
“Results from cohort C indicated a benefit with Opdivo regardless of the order of prior treatment with autologous hematopoietic stem cell transplantation and brentuximab vedotin, providing important insights as we continue researching the potential role Opdivo could provide for heavily pretreated classical Hodgkin lymphoma patients.”
The results were presented at the 10th International Symposium on Hodgkin Lymphoma (abstract T022). Abstracts from this meeting have been published in haematologica.
The CheckMate -205 trial is sponsored by Bristol-Myers Squibb.
About the trial
CheckMate -205 is a multi-cohort study in which investigators are evaluating the safety and efficacy of nivolumab in adults with cHL.
Cohort A included cHL patients who had received auto-HSCT and were BV-naïve (n=63). Cohort B included cHL patients who had received auto-HSCT followed by BV (n=80).
Cohort C included cHL patients who had received BV before and/or after auto-HSCT (n=100).
The study also includes a cohort D, which is currently enrolling and evaluating nivolumab in combination with chemotherapy in newly diagnosed, advanced-stage cHL patients who are treatment-naïve (n=50).
Patients enrolled in this trial have received nivolumab at 3 mg/kg intravenously every 2 weeks until disease progression or unacceptable toxicity. In cohort C, patients were also treated until investigator-assessed complete response (CR) lasting 1 year.
The study’s primary endpoint was ORR by IRRC assessment. Secondary and other exploratory endpoints included duration of response by IRRC assessment for CR rate and partial response rate, PFS by IRRC assessment, OS, and safety.
Response
The investigators presented results from cohort C (n=100), which included patients with cHL who had received BV before and/or after auto-HSCT.
At a median follow-up of 8.8 months, ORR per the IRRC was 73% (n=73) overall. The investigators said ORR was consistent across patient subgroups, regardless of the timing of prior BV relative to auto-HSCT.
The ORR was 70% (n=23) in patients who received BV only before auto-HSCT, 72% (n=41) in patients who received BV only after auto-HSCT, and 88% (n=7) in patients who received BV before and after auto-HSCT.
The CR rate was 17% (n=17) overall, 18.2% (n=6) in patients who received BV only before auto-HSCT, 12.3% (n=7) in patients who received BV only after auto-HSCT, and 38% (n=3) in patients who received BV before and after auto-HSCT.
Survival
The median PFS was 11.2 months (range, 8.5 months to not achieved) overall, 11.2 months (range, 8.5 months to not achieved) in patients who received BV only before auto-HSCT, 8.9 months (range, 8.3 months to not achieved) in patients who received BV only after auto-HSCT, and not achieved (range, 5.6 months to not achieved) in patients who received BV before and after auto-HSCT.
The 6-month OS was 93.9% overall, 97% in patients who received BV only before auto-HSCT, 91% in patients who received BV only after auto-HSCT, and 100% in patients who received BV before and after auto-HSCT.
Safety
Treatment-related adverse events (AEs) occurred in 68% of patients between the first dose and 30 days after the last dose of nivolumab. The most common treatment-related AEs were diarrhea, infusion-related reaction, and fatigue (11% each).
Grade 3/4 AEs occurred in 19% of patients. Serious treatment-related AEs were reported in 17% of patients, and treatment-related AEs leading to discontinuation occurred in 6% of patients.
At present, no treatment-related deaths have been reported. ![]()
Vector may make gene therapy safer

Photo courtesy of Washington
State University Spokane
Using modified foamy retroviral vectors to deliver gene therapy may reduce the risk of genotoxicity, according to research published in Scientific Reports.
These vectors have demonstrated promise in vitro, and researchers believe they could be used to treat X-linked severe combined immunodeficiency, thalassemia, and other diseases.
“We’ve started to translate this in collaboration with other scientists and medical doctors into the clinic,” said study author Grant Trobridge, PhD, of Washington State University Spokane.
Dr Trobridge and his colleagues said they decided to pursue foamy retroviral vectors as a delivery system for gene therapy because these vectors are less likely than gammaretroviral vectors or lentiviral vectors to activate nearby genes, including proto-oncogenes.
Still, the researchers altered the foamy retroviral vectors to change how they interact with target stem cells in an attempt to ensure the vectors would insert themselves into safer parts of the genome.
The team said they were able to retarget the foamy retroviral vectors away from genes and into satellite regions enriched for trimethylated histone H3 at lysine 9 by modifying the foamy virus Gag and Pol proteins.
These retargeted foamy retroviral vectors integrated near genes significantly less often than unmodified foamy retroviral vectors (P<0.001).
The researchers also noted that their retargeted foamy retroviral vectors can be produced at clinically relevant titers, and engineered target cells are not needed. Any target cell can be used by using alternate foamy helper plasmids during vector production. ![]()

Photo courtesy of Washington
State University Spokane
Using modified foamy retroviral vectors to deliver gene therapy may reduce the risk of genotoxicity, according to research published in Scientific Reports.
These vectors have demonstrated promise in vitro, and researchers believe they could be used to treat X-linked severe combined immunodeficiency, thalassemia, and other diseases.
“We’ve started to translate this in collaboration with other scientists and medical doctors into the clinic,” said study author Grant Trobridge, PhD, of Washington State University Spokane.
Dr Trobridge and his colleagues said they decided to pursue foamy retroviral vectors as a delivery system for gene therapy because these vectors are less likely than gammaretroviral vectors or lentiviral vectors to activate nearby genes, including proto-oncogenes.
Still, the researchers altered the foamy retroviral vectors to change how they interact with target stem cells in an attempt to ensure the vectors would insert themselves into safer parts of the genome.
The team said they were able to retarget the foamy retroviral vectors away from genes and into satellite regions enriched for trimethylated histone H3 at lysine 9 by modifying the foamy virus Gag and Pol proteins.
These retargeted foamy retroviral vectors integrated near genes significantly less often than unmodified foamy retroviral vectors (P<0.001).
The researchers also noted that their retargeted foamy retroviral vectors can be produced at clinically relevant titers, and engineered target cells are not needed. Any target cell can be used by using alternate foamy helper plasmids during vector production. ![]()

Photo courtesy of Washington
State University Spokane
Using modified foamy retroviral vectors to deliver gene therapy may reduce the risk of genotoxicity, according to research published in Scientific Reports.
These vectors have demonstrated promise in vitro, and researchers believe they could be used to treat X-linked severe combined immunodeficiency, thalassemia, and other diseases.
“We’ve started to translate this in collaboration with other scientists and medical doctors into the clinic,” said study author Grant Trobridge, PhD, of Washington State University Spokane.
Dr Trobridge and his colleagues said they decided to pursue foamy retroviral vectors as a delivery system for gene therapy because these vectors are less likely than gammaretroviral vectors or lentiviral vectors to activate nearby genes, including proto-oncogenes.
Still, the researchers altered the foamy retroviral vectors to change how they interact with target stem cells in an attempt to ensure the vectors would insert themselves into safer parts of the genome.
The team said they were able to retarget the foamy retroviral vectors away from genes and into satellite regions enriched for trimethylated histone H3 at lysine 9 by modifying the foamy virus Gag and Pol proteins.
These retargeted foamy retroviral vectors integrated near genes significantly less often than unmodified foamy retroviral vectors (P<0.001).
The researchers also noted that their retargeted foamy retroviral vectors can be produced at clinically relevant titers, and engineered target cells are not needed. Any target cell can be used by using alternate foamy helper plasmids during vector production. ![]()
Combating drug resistance in FLT3-mutated AML
with autophagosomes (green),
a process that happens during
mitophagy in cancer cells
treated with FLT3 inhibitor
Image from Besim Ogretmen
and Mohammed Dany/MUSC
Research published in Blood has revealed a mechanism that confers treatment resistance in FLT3-mutated acute myeloid leukemia (AML), as well as a drug that might overcome that resistance.
Researchers found that ceramide-dependent mitophagy plays a key role in drug-mediated AML cell death.
“Ceramide, a pro-cell-death lipid, kills cancer cells by causing them to eat their own mitochondria,” explained study author Besim Ogretmen, PhD, of the Medical University of South Carolina (MUSC) Hollings Cancer Center in Charleston, South Carolina.
AML cells with FLT3-ITD inhibit ceramide synthesis and thereby become resistant to cell death. FLT3 inhibitors have been developed to combat this resistance, but they’ve fallen short of expectations.
“Unfortunately, regardless of the inhibitor, the problem of resistance to FLT3-targeted therapy has persisted,” said study author Mohammed Dany, an MD/PhD student at MUSC.
However, Dany, Dr Ogretmen, and their colleagues were able to overcome this resistance with a synthetic ceramide analogue known as LCL-461.
In vitro, the drug reactivated mitophagy and killed AML cells that were resistant to treatment with the FLT3 inhibitor crenolanib.
In mice with crenolanib-resistant human AML xenografts, LCL-461 eliminated AML cells from the bone marrow.
A positively charged molecule, LCL-461 is attracted to the mitochondria of cancer cells, which become negatively charged through the Warburg effect. The researchers said this limits off-target effects that can occur with less specific inhibitors of FLT3 signaling.
Furthermore, Dr Ogretmen’s lab has tested the safety of LCL-461 in previous studies and reported that it had no major side effects at therapeutically active doses.
Dr Ogretmen and his colleagues’ next step is to perform large animal studies with LCL-461.
“We are very excited about this,” Dr Ogretmen said. “Head and neck cancers also respond to this drug very well. What we are trying to do is really cure cancer one disease at a time, and we are digging and digging to understand the mechanisms of how these cancer cells escape therapeutic interventions so that we can find mechanism-based therapeutics to have more tools for treatment.”
LCL-461 was developed at MUSC. The MUSC Foundation for Research Development has patented the drug and licensed it to Charleston-based startup SphingoGene, Inc. ![]()

with autophagosomes (green),
a process that happens during
mitophagy in cancer cells
treated with FLT3 inhibitor
Image from Besim Ogretmen
and Mohammed Dany/MUSC
Research published in Blood has revealed a mechanism that confers treatment resistance in FLT3-mutated acute myeloid leukemia (AML), as well as a drug that might overcome that resistance.
Researchers found that ceramide-dependent mitophagy plays a key role in drug-mediated AML cell death.
“Ceramide, a pro-cell-death lipid, kills cancer cells by causing them to eat their own mitochondria,” explained study author Besim Ogretmen, PhD, of the Medical University of South Carolina (MUSC) Hollings Cancer Center in Charleston, South Carolina.
AML cells with FLT3-ITD inhibit ceramide synthesis and thereby become resistant to cell death. FLT3 inhibitors have been developed to combat this resistance, but they’ve fallen short of expectations.
“Unfortunately, regardless of the inhibitor, the problem of resistance to FLT3-targeted therapy has persisted,” said study author Mohammed Dany, an MD/PhD student at MUSC.
However, Dany, Dr Ogretmen, and their colleagues were able to overcome this resistance with a synthetic ceramide analogue known as LCL-461.
In vitro, the drug reactivated mitophagy and killed AML cells that were resistant to treatment with the FLT3 inhibitor crenolanib.
In mice with crenolanib-resistant human AML xenografts, LCL-461 eliminated AML cells from the bone marrow.
A positively charged molecule, LCL-461 is attracted to the mitochondria of cancer cells, which become negatively charged through the Warburg effect. The researchers said this limits off-target effects that can occur with less specific inhibitors of FLT3 signaling.
Furthermore, Dr Ogretmen’s lab has tested the safety of LCL-461 in previous studies and reported that it had no major side effects at therapeutically active doses.
Dr Ogretmen and his colleagues’ next step is to perform large animal studies with LCL-461.
“We are very excited about this,” Dr Ogretmen said. “Head and neck cancers also respond to this drug very well. What we are trying to do is really cure cancer one disease at a time, and we are digging and digging to understand the mechanisms of how these cancer cells escape therapeutic interventions so that we can find mechanism-based therapeutics to have more tools for treatment.”
LCL-461 was developed at MUSC. The MUSC Foundation for Research Development has patented the drug and licensed it to Charleston-based startup SphingoGene, Inc. ![]()

with autophagosomes (green),
a process that happens during
mitophagy in cancer cells
treated with FLT3 inhibitor
Image from Besim Ogretmen
and Mohammed Dany/MUSC
Research published in Blood has revealed a mechanism that confers treatment resistance in FLT3-mutated acute myeloid leukemia (AML), as well as a drug that might overcome that resistance.
Researchers found that ceramide-dependent mitophagy plays a key role in drug-mediated AML cell death.
“Ceramide, a pro-cell-death lipid, kills cancer cells by causing them to eat their own mitochondria,” explained study author Besim Ogretmen, PhD, of the Medical University of South Carolina (MUSC) Hollings Cancer Center in Charleston, South Carolina.
AML cells with FLT3-ITD inhibit ceramide synthesis and thereby become resistant to cell death. FLT3 inhibitors have been developed to combat this resistance, but they’ve fallen short of expectations.
“Unfortunately, regardless of the inhibitor, the problem of resistance to FLT3-targeted therapy has persisted,” said study author Mohammed Dany, an MD/PhD student at MUSC.
However, Dany, Dr Ogretmen, and their colleagues were able to overcome this resistance with a synthetic ceramide analogue known as LCL-461.
In vitro, the drug reactivated mitophagy and killed AML cells that were resistant to treatment with the FLT3 inhibitor crenolanib.
In mice with crenolanib-resistant human AML xenografts, LCL-461 eliminated AML cells from the bone marrow.
A positively charged molecule, LCL-461 is attracted to the mitochondria of cancer cells, which become negatively charged through the Warburg effect. The researchers said this limits off-target effects that can occur with less specific inhibitors of FLT3 signaling.
Furthermore, Dr Ogretmen’s lab has tested the safety of LCL-461 in previous studies and reported that it had no major side effects at therapeutically active doses.
Dr Ogretmen and his colleagues’ next step is to perform large animal studies with LCL-461.
“We are very excited about this,” Dr Ogretmen said. “Head and neck cancers also respond to this drug very well. What we are trying to do is really cure cancer one disease at a time, and we are digging and digging to understand the mechanisms of how these cancer cells escape therapeutic interventions so that we can find mechanism-based therapeutics to have more tools for treatment.”
LCL-461 was developed at MUSC. The MUSC Foundation for Research Development has patented the drug and licensed it to Charleston-based startup SphingoGene, Inc. ![]()
Team develops multicolor electron microscopy
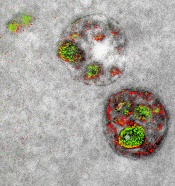
of endosomal uptake
of peptide proteins
Image by Adams et al/
Cell Chemical Biology 2016
Researchers say they have found a way to image specific cellular components using multicolor electron microscopy.
With their method, which the team worked on for nearly 15 years, up to 3 colors at a time (green, red, or yellow) can be used in an image.
A detector on the microscope captures electrons lost from metal ions painted over the specimen and records the metal’s energy loss signature as a color.
A technician must add the ionized metals one at a time and then lay the full color map over the still microscopy image.
The researchers described this method in detail in Cell Chemical Biology.
“This method has many potential applications in biology,” said study author Stephen Adams, PhD, of the University of California, San Diego in La Jolla.
“In the paper, we demonstrate how it can distinguish cellular compartments or track proteins and tag cells.”
For the multicolor effect to work, the researchers needed metal complexes that are stable enough to withstand application (meaning they don’t quickly deteriorate and blur the image) and have a distinct electron energy loss signature.
The researchers used ionized lanthanum (La), cerium (Ce), and praseodymium (Pr)—all metals in the lanthanide family. Each metal complex was laid down sequentially as a precipitate onto the specimen as it sat in the microscope.
“One challenge that kept us from publishing this much earlier, because we had the chemistry and we had an instrument that worked about 4 years ago, was we needed a way to deposit the metal compounds sequentially,” said study author Mark Ellisman, PhD, of the University of California, San Diego.
“We spent an awful lot of time trying to figure out how to deposit one of the lanthanides and then clear it so that it didn’t react when we deposited a second signal on the first site.”
Once the application process had been established, the researchers illustrated the power of multicolor electron microscopy by visualizing 2 brain cells sharing a single synapse. They also showed peptides entering through a cell membrane.
The researchers said there is more chemistry to be done to perfect the metal ion application process as well as produce images with more colors. There may also be ways to increase the amount of metal ions that can be deposited, which could help with resolution.
Many in the biochemical community should be able to begin using this technique right away, as it takes advantage of tools that are already found in laboratories.
This study is one of the last that Roger Tsien, PhD, who won a 2008 Nobel Prize in Chemistry for the discovery and application of green fluorescent protein to biochemical imaging, saw accepted by a journal before his death last August.
He did the first experiments to develop the chemical compounds needed for the multicolor imaging method nearly 15 years ago.
“One theme that has gone through all of Roger’s work is the desire to peer more closely into the workings of the cell,” Dr Adams said.
“With all of the fluorescence techniques that he’s introduced, he was able to do that in live cells and make action movies of them in vivid colors. But he always wanted to look closer, and now he’s left the beginnings for a method where we can add colors to electron microscopy.”
“This is clearly an example of Roger’s brilliance at chemistry and how he saw that, if we could do this, we would be able to enjoy the advantages of electron microscopy,” Dr Ellisman added.
“The biggest advantage of electron microscopy that we saw is that you have weak contrasts by the nature of the way that staining works, so color-specific labels give context to all of the rich information in the scene of which molecules are operating.” ![]()

of endosomal uptake
of peptide proteins
Image by Adams et al/
Cell Chemical Biology 2016
Researchers say they have found a way to image specific cellular components using multicolor electron microscopy.
With their method, which the team worked on for nearly 15 years, up to 3 colors at a time (green, red, or yellow) can be used in an image.
A detector on the microscope captures electrons lost from metal ions painted over the specimen and records the metal’s energy loss signature as a color.
A technician must add the ionized metals one at a time and then lay the full color map over the still microscopy image.
The researchers described this method in detail in Cell Chemical Biology.
“This method has many potential applications in biology,” said study author Stephen Adams, PhD, of the University of California, San Diego in La Jolla.
“In the paper, we demonstrate how it can distinguish cellular compartments or track proteins and tag cells.”
For the multicolor effect to work, the researchers needed metal complexes that are stable enough to withstand application (meaning they don’t quickly deteriorate and blur the image) and have a distinct electron energy loss signature.
The researchers used ionized lanthanum (La), cerium (Ce), and praseodymium (Pr)—all metals in the lanthanide family. Each metal complex was laid down sequentially as a precipitate onto the specimen as it sat in the microscope.
“One challenge that kept us from publishing this much earlier, because we had the chemistry and we had an instrument that worked about 4 years ago, was we needed a way to deposit the metal compounds sequentially,” said study author Mark Ellisman, PhD, of the University of California, San Diego.
“We spent an awful lot of time trying to figure out how to deposit one of the lanthanides and then clear it so that it didn’t react when we deposited a second signal on the first site.”
Once the application process had been established, the researchers illustrated the power of multicolor electron microscopy by visualizing 2 brain cells sharing a single synapse. They also showed peptides entering through a cell membrane.
The researchers said there is more chemistry to be done to perfect the metal ion application process as well as produce images with more colors. There may also be ways to increase the amount of metal ions that can be deposited, which could help with resolution.
Many in the biochemical community should be able to begin using this technique right away, as it takes advantage of tools that are already found in laboratories.
This study is one of the last that Roger Tsien, PhD, who won a 2008 Nobel Prize in Chemistry for the discovery and application of green fluorescent protein to biochemical imaging, saw accepted by a journal before his death last August.
He did the first experiments to develop the chemical compounds needed for the multicolor imaging method nearly 15 years ago.
“One theme that has gone through all of Roger’s work is the desire to peer more closely into the workings of the cell,” Dr Adams said.
“With all of the fluorescence techniques that he’s introduced, he was able to do that in live cells and make action movies of them in vivid colors. But he always wanted to look closer, and now he’s left the beginnings for a method where we can add colors to electron microscopy.”
“This is clearly an example of Roger’s brilliance at chemistry and how he saw that, if we could do this, we would be able to enjoy the advantages of electron microscopy,” Dr Ellisman added.
“The biggest advantage of electron microscopy that we saw is that you have weak contrasts by the nature of the way that staining works, so color-specific labels give context to all of the rich information in the scene of which molecules are operating.” ![]()

of endosomal uptake
of peptide proteins
Image by Adams et al/
Cell Chemical Biology 2016
Researchers say they have found a way to image specific cellular components using multicolor electron microscopy.
With their method, which the team worked on for nearly 15 years, up to 3 colors at a time (green, red, or yellow) can be used in an image.
A detector on the microscope captures electrons lost from metal ions painted over the specimen and records the metal’s energy loss signature as a color.
A technician must add the ionized metals one at a time and then lay the full color map over the still microscopy image.
The researchers described this method in detail in Cell Chemical Biology.
“This method has many potential applications in biology,” said study author Stephen Adams, PhD, of the University of California, San Diego in La Jolla.
“In the paper, we demonstrate how it can distinguish cellular compartments or track proteins and tag cells.”
For the multicolor effect to work, the researchers needed metal complexes that are stable enough to withstand application (meaning they don’t quickly deteriorate and blur the image) and have a distinct electron energy loss signature.
The researchers used ionized lanthanum (La), cerium (Ce), and praseodymium (Pr)—all metals in the lanthanide family. Each metal complex was laid down sequentially as a precipitate onto the specimen as it sat in the microscope.
“One challenge that kept us from publishing this much earlier, because we had the chemistry and we had an instrument that worked about 4 years ago, was we needed a way to deposit the metal compounds sequentially,” said study author Mark Ellisman, PhD, of the University of California, San Diego.
“We spent an awful lot of time trying to figure out how to deposit one of the lanthanides and then clear it so that it didn’t react when we deposited a second signal on the first site.”
Once the application process had been established, the researchers illustrated the power of multicolor electron microscopy by visualizing 2 brain cells sharing a single synapse. They also showed peptides entering through a cell membrane.
The researchers said there is more chemistry to be done to perfect the metal ion application process as well as produce images with more colors. There may also be ways to increase the amount of metal ions that can be deposited, which could help with resolution.
Many in the biochemical community should be able to begin using this technique right away, as it takes advantage of tools that are already found in laboratories.
This study is one of the last that Roger Tsien, PhD, who won a 2008 Nobel Prize in Chemistry for the discovery and application of green fluorescent protein to biochemical imaging, saw accepted by a journal before his death last August.
He did the first experiments to develop the chemical compounds needed for the multicolor imaging method nearly 15 years ago.
“One theme that has gone through all of Roger’s work is the desire to peer more closely into the workings of the cell,” Dr Adams said.
“With all of the fluorescence techniques that he’s introduced, he was able to do that in live cells and make action movies of them in vivid colors. But he always wanted to look closer, and now he’s left the beginnings for a method where we can add colors to electron microscopy.”
“This is clearly an example of Roger’s brilliance at chemistry and how he saw that, if we could do this, we would be able to enjoy the advantages of electron microscopy,” Dr Ellisman added.
“The biggest advantage of electron microscopy that we saw is that you have weak contrasts by the nature of the way that staining works, so color-specific labels give context to all of the rich information in the scene of which molecules are operating.”
Combine flow and HTS for sensitive MRD detection in CLL, speaker says

laser beam
Photo courtesy of NIH
NEW YORK—Combining 2 technologies—flow cytometry and high-throughput sequencing (HTS)—produces a very sensitive approach to detecting minimal residual disease (MRD) in chronic lymphocytic leukemia (CLL), according to a speaker at Lymphoma & Myeloma 2016.
The approach is both reproducible and widely accessible, added the speaker, Peter Hillmen, MB ChB, PhD, of St James’s University Hospital in Leeds, UK.
“PCR [polymerase chain reaction] is more sensitive than flow cytometry,” he noted, “but it is probably not necessary to assess response by that criteria.”
Features to consider when choosing a technology for detecting MRD include sensitivity, specificity for the patient and/or the disease, applicability to all patients or to an individual, the platform—flow cytometry, conventional molecular PCR, or next-generation sequencing—and which tissue to use, blood or bone marrow.
In the era of immunotherapy, Dr Hillmen said, assessment should be performed at least 2 months after completion of immunotherapy to get a reliable assessment of MRD, particularly after treatment with alemtuzumab, rituximab, and other antibodies targeting CLL.
History of MRD analysis
The definition of MRD hasn’t changed since 2008, when the International Workshop on CLL updated National Cancer Institute guidelines. It is still 1 single cell in 10,000 (10-4) leukocytes, regardless of the tissue used.
Prior to the mid-1990s, there were limited options for assessing MRD, Dr Hillmen said.
Based on the profound remissions patients experienced with alemtuzumab, investigators began to develop assays to assess MRD.
“[W]e started standardizing these assays around 2007,” Dr Hillmen said, and a standardized assay was used prospectively in clinical trials beginning in 2012.
Technologies
Several technologies can be used to assess MRD.
Flow cytometry using 6 colors or 8 colors—to simplify the assay and to make it more sensitive—is a multiparameter assessment of CLL phenotype that is not clonality-based.
Allele-specific oligonucleotide PCR (ASO-PCR) is laborious to perform, Dr Hillmen said, but it’s very sensitive.
“[I]t probably shouldn’t be considered as an MRD test,” he said, since it uses patient-specific primers, not consensus primers.
HTS provides an increasing amount of information on B-cell sequences and enumeration of the CLL-specific immunoglobulin gene, “and I would move it towards being approved as a regulatory endpoint,” Dr Hillmen asserted.
Flow cytometry and HTS
A consensus document by the European Research Initiative on CLL (ERIC) identified and validated a flow cytometric approach to MRD assessment in parallel with HTS.
According to the ERIC investigators, flow cytometry had to utilize a core panel of 6 antigens used by most labs—CD19, CD20, CD5, CD43, CD79b, and CD81. And the markers used had to quantitate cells to a level of 0.01% (10-4).
Assays had to be independent and compatible with older, established therapies as well as newer treatments.
For example, 6-color flow had to be effective with fludarabine, cyclophosphamide, and rituximab (FCR) regimens, as well as effective with the novel agents ibrutinib and venetoclax (ABT-199).
Compared to PCR, multiparameter flow cytometry is more convenient, Dr Hillmen noted.
And while PCR is more sensitive than flow cytometry (sensitive to 10-5 to 10-6), it is more difficult to apply to large clinical trials because the assay must be validated for each patient.
The investigators validated the flow cytometric approach in 450 patients on FCR-type therapy enrolled in the ADMIRE and ARCTIC trials.
They assessed MRD in patients’ bone marrow 3 months after the last course of treatment and presented the data at EHA 2015 (Rawstron AC, abstract S794).
They found that all patients who were MRD negative, including 9 patients with partial responses, achieved a significantly better progression-free survival (PFS) than patients who had achieved a complete response but were still MRD positive.
The parallel analysis of HTS showed good concordance with flow cytometry at the 0.01% (10-4) level.
Peripheral blood or bone marrow?
“[T]he blood is a tissue which can be used, but it’s certainly not as sensitive as the bone marrow,” Dr Hillmen said. “And depending upon what we are using MRD for, the marrow is probably a better tissue, with some exceptions.”
Data from the ADMIRE and ARCTIC trials confirmed that 177 patients on FCR-based therapy who were negative in the bone marrow were always negative in the blood. However, a quarter of patients negative in the blood were positive in the bone marrow.
Investigators followed the same patients on FCR-based therapy for 3 years and found no difference in outcome in terms of PFS for patients negative in peripheral blood and positive in the bone marrow (PB-/BM+) and those negative in peripheral blood and negative in the bone marrow (PB-/BM-) (Rawstron abstract S794).
But for patients on alemtuzumab, with the same analysis, those who were PB-/BM+ did less well and had similar PFS to those who were PB+/BM+.
And at a follow-up of 4 years or longer, patients on FCR-based therapy who were PB-/BM- had superior outcomes than those who were PB-/BM+.
“So as a predictive marker, the bone marrow is a better tissue to look at, but peripheral blood negativity also can predict with FCR but not with agents such as alemtuzumab,” Dr Hillmen summarized.
Prognostic value of MRD assessment
Multivariate analysis of a 10-year follow-up of 133 CLL patients revealed that MRD level and adverse cytogenetics were the only significant parameters in terms of PFS.
And in terms of overall survival (OS), MRD level, prior treatment, Binet stage, and age were significant.
Sixty-seven of these patients had been treated with chemoimmunotherapy, 31 with single-agent chemotherapy, 7 had autologous stem cell transplants, and 28 had prior exposure to alemtuzumab.
In terms of survival beyond 10 or 15 years, previously untreated patients who were MRD negative after their first therapy had significantly better PFS and OS than previously treated patients who were MRD negative and patients with or without prior treatment who were MRD positive (P<0.001).
“Consistently, MRD, regardless of therapy, is the most important prognostic marker,” Dr Hillmen said.
Data from MD Anderson Cancer Center showed that 75% of patients treated first-line with FCR who achieved a complete response were MRD negative.
And patients who achieved MRD negativity had significantly better PFS (P<0.001) and OS (P=0.006) than patients who remained MRD positive.
In the ADMIRE and ARCTIC trials mentioned earlier, patients who achieved MRD negativity in the marrow at 3 months post-therapy also had significantly better PFS (P<0.0001) and OS (P=0.0002) than those who were MRD positive.
For every log of positivity, Dr Hillmen said, patients have a worse survival. Conversely, for every log reduction in MRD level, there is a 33% reduction in risk for disease progression.
“MRD is probably the most important prognostic marker we have,” he said. “We need to look at MRD levels with novel agents and use it to define duration of therapy, maybe use it to define additional therapy if patients are stalled in their response.”

laser beam
Photo courtesy of NIH
NEW YORK—Combining 2 technologies—flow cytometry and high-throughput sequencing (HTS)—produces a very sensitive approach to detecting minimal residual disease (MRD) in chronic lymphocytic leukemia (CLL), according to a speaker at Lymphoma & Myeloma 2016.
The approach is both reproducible and widely accessible, added the speaker, Peter Hillmen, MB ChB, PhD, of St James’s University Hospital in Leeds, UK.
“PCR [polymerase chain reaction] is more sensitive than flow cytometry,” he noted, “but it is probably not necessary to assess response by that criteria.”
Features to consider when choosing a technology for detecting MRD include sensitivity, specificity for the patient and/or the disease, applicability to all patients or to an individual, the platform—flow cytometry, conventional molecular PCR, or next-generation sequencing—and which tissue to use, blood or bone marrow.
In the era of immunotherapy, Dr Hillmen said, assessment should be performed at least 2 months after completion of immunotherapy to get a reliable assessment of MRD, particularly after treatment with alemtuzumab, rituximab, and other antibodies targeting CLL.
History of MRD analysis
The definition of MRD hasn’t changed since 2008, when the International Workshop on CLL updated National Cancer Institute guidelines. It is still 1 single cell in 10,000 (10-4) leukocytes, regardless of the tissue used.
Prior to the mid-1990s, there were limited options for assessing MRD, Dr Hillmen said.
Based on the profound remissions patients experienced with alemtuzumab, investigators began to develop assays to assess MRD.
“[W]e started standardizing these assays around 2007,” Dr Hillmen said, and a standardized assay was used prospectively in clinical trials beginning in 2012.
Technologies
Several technologies can be used to assess MRD.
Flow cytometry using 6 colors or 8 colors—to simplify the assay and to make it more sensitive—is a multiparameter assessment of CLL phenotype that is not clonality-based.
Allele-specific oligonucleotide PCR (ASO-PCR) is laborious to perform, Dr Hillmen said, but it’s very sensitive.
“[I]t probably shouldn’t be considered as an MRD test,” he said, since it uses patient-specific primers, not consensus primers.
HTS provides an increasing amount of information on B-cell sequences and enumeration of the CLL-specific immunoglobulin gene, “and I would move it towards being approved as a regulatory endpoint,” Dr Hillmen asserted.
Flow cytometry and HTS
A consensus document by the European Research Initiative on CLL (ERIC) identified and validated a flow cytometric approach to MRD assessment in parallel with HTS.
According to the ERIC investigators, flow cytometry had to utilize a core panel of 6 antigens used by most labs—CD19, CD20, CD5, CD43, CD79b, and CD81. And the markers used had to quantitate cells to a level of 0.01% (10-4).
Assays had to be independent and compatible with older, established therapies as well as newer treatments.
For example, 6-color flow had to be effective with fludarabine, cyclophosphamide, and rituximab (FCR) regimens, as well as effective with the novel agents ibrutinib and venetoclax (ABT-199).
Compared to PCR, multiparameter flow cytometry is more convenient, Dr Hillmen noted.
And while PCR is more sensitive than flow cytometry (sensitive to 10-5 to 10-6), it is more difficult to apply to large clinical trials because the assay must be validated for each patient.
The investigators validated the flow cytometric approach in 450 patients on FCR-type therapy enrolled in the ADMIRE and ARCTIC trials.
They assessed MRD in patients’ bone marrow 3 months after the last course of treatment and presented the data at EHA 2015 (Rawstron AC, abstract S794).
They found that all patients who were MRD negative, including 9 patients with partial responses, achieved a significantly better progression-free survival (PFS) than patients who had achieved a complete response but were still MRD positive.
The parallel analysis of HTS showed good concordance with flow cytometry at the 0.01% (10-4) level.
Peripheral blood or bone marrow?
“[T]he blood is a tissue which can be used, but it’s certainly not as sensitive as the bone marrow,” Dr Hillmen said. “And depending upon what we are using MRD for, the marrow is probably a better tissue, with some exceptions.”
Data from the ADMIRE and ARCTIC trials confirmed that 177 patients on FCR-based therapy who were negative in the bone marrow were always negative in the blood. However, a quarter of patients negative in the blood were positive in the bone marrow.
Investigators followed the same patients on FCR-based therapy for 3 years and found no difference in outcome in terms of PFS for patients negative in peripheral blood and positive in the bone marrow (PB-/BM+) and those negative in peripheral blood and negative in the bone marrow (PB-/BM-) (Rawstron abstract S794).
But for patients on alemtuzumab, with the same analysis, those who were PB-/BM+ did less well and had similar PFS to those who were PB+/BM+.
And at a follow-up of 4 years or longer, patients on FCR-based therapy who were PB-/BM- had superior outcomes than those who were PB-/BM+.
“So as a predictive marker, the bone marrow is a better tissue to look at, but peripheral blood negativity also can predict with FCR but not with agents such as alemtuzumab,” Dr Hillmen summarized.
Prognostic value of MRD assessment
Multivariate analysis of a 10-year follow-up of 133 CLL patients revealed that MRD level and adverse cytogenetics were the only significant parameters in terms of PFS.
And in terms of overall survival (OS), MRD level, prior treatment, Binet stage, and age were significant.
Sixty-seven of these patients had been treated with chemoimmunotherapy, 31 with single-agent chemotherapy, 7 had autologous stem cell transplants, and 28 had prior exposure to alemtuzumab.
In terms of survival beyond 10 or 15 years, previously untreated patients who were MRD negative after their first therapy had significantly better PFS and OS than previously treated patients who were MRD negative and patients with or without prior treatment who were MRD positive (P<0.001).
“Consistently, MRD, regardless of therapy, is the most important prognostic marker,” Dr Hillmen said.
Data from MD Anderson Cancer Center showed that 75% of patients treated first-line with FCR who achieved a complete response were MRD negative.
And patients who achieved MRD negativity had significantly better PFS (P<0.001) and OS (P=0.006) than patients who remained MRD positive.
In the ADMIRE and ARCTIC trials mentioned earlier, patients who achieved MRD negativity in the marrow at 3 months post-therapy also had significantly better PFS (P<0.0001) and OS (P=0.0002) than those who were MRD positive.
For every log of positivity, Dr Hillmen said, patients have a worse survival. Conversely, for every log reduction in MRD level, there is a 33% reduction in risk for disease progression.
“MRD is probably the most important prognostic marker we have,” he said. “We need to look at MRD levels with novel agents and use it to define duration of therapy, maybe use it to define additional therapy if patients are stalled in their response.”

laser beam
Photo courtesy of NIH
NEW YORK—Combining 2 technologies—flow cytometry and high-throughput sequencing (HTS)—produces a very sensitive approach to detecting minimal residual disease (MRD) in chronic lymphocytic leukemia (CLL), according to a speaker at Lymphoma & Myeloma 2016.
The approach is both reproducible and widely accessible, added the speaker, Peter Hillmen, MB ChB, PhD, of St James’s University Hospital in Leeds, UK.
“PCR [polymerase chain reaction] is more sensitive than flow cytometry,” he noted, “but it is probably not necessary to assess response by that criteria.”
Features to consider when choosing a technology for detecting MRD include sensitivity, specificity for the patient and/or the disease, applicability to all patients or to an individual, the platform—flow cytometry, conventional molecular PCR, or next-generation sequencing—and which tissue to use, blood or bone marrow.
In the era of immunotherapy, Dr Hillmen said, assessment should be performed at least 2 months after completion of immunotherapy to get a reliable assessment of MRD, particularly after treatment with alemtuzumab, rituximab, and other antibodies targeting CLL.
History of MRD analysis
The definition of MRD hasn’t changed since 2008, when the International Workshop on CLL updated National Cancer Institute guidelines. It is still 1 single cell in 10,000 (10-4) leukocytes, regardless of the tissue used.
Prior to the mid-1990s, there were limited options for assessing MRD, Dr Hillmen said.
Based on the profound remissions patients experienced with alemtuzumab, investigators began to develop assays to assess MRD.
“[W]e started standardizing these assays around 2007,” Dr Hillmen said, and a standardized assay was used prospectively in clinical trials beginning in 2012.
Technologies
Several technologies can be used to assess MRD.
Flow cytometry using 6 colors or 8 colors—to simplify the assay and to make it more sensitive—is a multiparameter assessment of CLL phenotype that is not clonality-based.
Allele-specific oligonucleotide PCR (ASO-PCR) is laborious to perform, Dr Hillmen said, but it’s very sensitive.
“[I]t probably shouldn’t be considered as an MRD test,” he said, since it uses patient-specific primers, not consensus primers.
HTS provides an increasing amount of information on B-cell sequences and enumeration of the CLL-specific immunoglobulin gene, “and I would move it towards being approved as a regulatory endpoint,” Dr Hillmen asserted.
Flow cytometry and HTS
A consensus document by the European Research Initiative on CLL (ERIC) identified and validated a flow cytometric approach to MRD assessment in parallel with HTS.
According to the ERIC investigators, flow cytometry had to utilize a core panel of 6 antigens used by most labs—CD19, CD20, CD5, CD43, CD79b, and CD81. And the markers used had to quantitate cells to a level of 0.01% (10-4).
Assays had to be independent and compatible with older, established therapies as well as newer treatments.
For example, 6-color flow had to be effective with fludarabine, cyclophosphamide, and rituximab (FCR) regimens, as well as effective with the novel agents ibrutinib and venetoclax (ABT-199).
Compared to PCR, multiparameter flow cytometry is more convenient, Dr Hillmen noted.
And while PCR is more sensitive than flow cytometry (sensitive to 10-5 to 10-6), it is more difficult to apply to large clinical trials because the assay must be validated for each patient.
The investigators validated the flow cytometric approach in 450 patients on FCR-type therapy enrolled in the ADMIRE and ARCTIC trials.
They assessed MRD in patients’ bone marrow 3 months after the last course of treatment and presented the data at EHA 2015 (Rawstron AC, abstract S794).
They found that all patients who were MRD negative, including 9 patients with partial responses, achieved a significantly better progression-free survival (PFS) than patients who had achieved a complete response but were still MRD positive.
The parallel analysis of HTS showed good concordance with flow cytometry at the 0.01% (10-4) level.
Peripheral blood or bone marrow?
“[T]he blood is a tissue which can be used, but it’s certainly not as sensitive as the bone marrow,” Dr Hillmen said. “And depending upon what we are using MRD for, the marrow is probably a better tissue, with some exceptions.”
Data from the ADMIRE and ARCTIC trials confirmed that 177 patients on FCR-based therapy who were negative in the bone marrow were always negative in the blood. However, a quarter of patients negative in the blood were positive in the bone marrow.
Investigators followed the same patients on FCR-based therapy for 3 years and found no difference in outcome in terms of PFS for patients negative in peripheral blood and positive in the bone marrow (PB-/BM+) and those negative in peripheral blood and negative in the bone marrow (PB-/BM-) (Rawstron abstract S794).
But for patients on alemtuzumab, with the same analysis, those who were PB-/BM+ did less well and had similar PFS to those who were PB+/BM+.
And at a follow-up of 4 years or longer, patients on FCR-based therapy who were PB-/BM- had superior outcomes than those who were PB-/BM+.
“So as a predictive marker, the bone marrow is a better tissue to look at, but peripheral blood negativity also can predict with FCR but not with agents such as alemtuzumab,” Dr Hillmen summarized.
Prognostic value of MRD assessment
Multivariate analysis of a 10-year follow-up of 133 CLL patients revealed that MRD level and adverse cytogenetics were the only significant parameters in terms of PFS.
And in terms of overall survival (OS), MRD level, prior treatment, Binet stage, and age were significant.
Sixty-seven of these patients had been treated with chemoimmunotherapy, 31 with single-agent chemotherapy, 7 had autologous stem cell transplants, and 28 had prior exposure to alemtuzumab.
In terms of survival beyond 10 or 15 years, previously untreated patients who were MRD negative after their first therapy had significantly better PFS and OS than previously treated patients who were MRD negative and patients with or without prior treatment who were MRD positive (P<0.001).
“Consistently, MRD, regardless of therapy, is the most important prognostic marker,” Dr Hillmen said.
Data from MD Anderson Cancer Center showed that 75% of patients treated first-line with FCR who achieved a complete response were MRD negative.
And patients who achieved MRD negativity had significantly better PFS (P<0.001) and OS (P=0.006) than patients who remained MRD positive.
In the ADMIRE and ARCTIC trials mentioned earlier, patients who achieved MRD negativity in the marrow at 3 months post-therapy also had significantly better PFS (P<0.0001) and OS (P=0.0002) than those who were MRD positive.
For every log of positivity, Dr Hillmen said, patients have a worse survival. Conversely, for every log reduction in MRD level, there is a 33% reduction in risk for disease progression.
“MRD is probably the most important prognostic marker we have,” he said. “We need to look at MRD levels with novel agents and use it to define duration of therapy, maybe use it to define additional therapy if patients are stalled in their response.”
Studies reveal markers of malaria resistance
Plasmodium falciparum
Image from CDC/Mae Melvin
Two studies have revealed genetic markers associated with resistance to piperaquine, one of the most commonly used malaria drugs in Southeast Asia.
Investigators found that Plasmodium falciparum parasites were less sensitive to piperaquine if they had the exo-E415G variant or multiple copies of the plasmepsin 2 and plasmepsin 3 genes.
Furthermore, the presence of these markers could identify which malaria patients would fail piperaquine treatment.
However, investigators said more research is needed to establish whether plasmepsin gene amplification and the exo-E415G variant actually cause piperaquine resistance and to explore whether this association extends to other parts of the world.
The 2 groups of investigators reported the findings of their studies in The Lancet Infectious Diseases.
About resistance
The combination of artemisinin and piperaquine (dihydroartemisinin-piperaquine) is a frontline treatment for malaria in Southeast Asia.
However, in recent years, the emergence of resistance to these drugs in Cambodia means that up to 60% of patients fail treatment in several provinces. There are fears that resistance could spread to other regions where malaria is endemic.
Genome-wide association studies have been conducted to identify genetic variations among P falciparum parasites that can be linked to variations in drug resistance.
Recent studies have revealed genetic markers linked to resistance to artemisinin (K13) and mefloquine (Pfmdr1), but, until now, markers for resistance to other combination drugs had not been identified.
Plasmepsin genes
In the first study, Didier Ménard, PhD, of the Institut Pasteur in Phnom Penh, Cambodia, and his colleagues found that amplification of plasmepsin genes in the P falciparum genome was linked to piperaquine resistance.
The investigators first analyzed artemisinin-resistant, Cambodian P falciparum parasite lines, comparing the exomes of lines that were piperaquine-susceptible and those that were piperaquine-resistant.
The team said this revealed an increased copy number of the plasmepsin 2-plasmepsin 3 gene cluster as a putative genetic signature associated with piperaquine resistance.
The investigators then confirmed their findings in a set of 725 P falciparum parasites collected from patients across Cambodia since 2009.
Individuals infected with parasites with multiple copies of the plasmepsin 2 gene were, on average, 20.4 times more likely to experience dihydroartemisinin-piperaquine treatment failure.
Furthermore, the geographical distribution and rise in the proportion of parasites with multiple copies of the plasmepsin 2 and 3 genes in Cambodia corresponded with areas that had reported increases in dihydroartemisinin-piperaquine treatment failures in recent years.
“We now know that amplification of the plasmepsin 2 and plasmepsin 3 genes is an important factor in determining how well piperaquine kills malaria parasites,” Dr Ménard said. “A genetic toolkit combining this new marker with markers of artemisinin and mefloquine resistance could be used to track the spread of resistance and provide timely information for containment policies.”
“Piperaquine resistance, although currently confined to Cambodia, is a major concern, because patients suffering malaria are almost untreatable. At present, alternative treatments are scarce, and the reduced cure rates lead to prolonged parasite carriage, increasing the potential of transmitting the resistant parasites and endangering efforts to control and eliminate malaria.”
Exo-E415G variant
The second study was conducted by Rick Fairhurst, MD, of the National Institutes of Health in Bethesda, Maryland, and his colleagues.
The investigators analyzed 486 P falciparum parasites collected between July 9, 2010, and December 31, 2013, from 3 different provinces in Cambodia where artemisinin resistance and dihydroartemisinin-piperaquine treatment failure are common (Pursat), emerging (Preah Vihear), or uncommon (Ratanakiri).
The investigators exposed the parasites to therapeutic levels of piperaquine and measured how well each strain grew. At the same time, they sequenced the entire genome of each parasite.
Parasites were more likely to survive exposure to piperaquine if they had 2 or more copies of the plasmepsin 2 and plasmepsin 3 genes, as well as the exo-E415G variant on chromosome 13.
Similarly, in a further analysis of 133 patient samples, the investigators found that individuals infected with parasites carrying multiple copies of the plasmepsin genes were much more likely to fail treatment with dihydroartemisinin-piperaquine, as were individuals who had parasites with the exo-E415G variant.
The team also noted that the prevalence of exo-E415G and plasmepsin 2-3 markers has risen substantially in recent years in Pursat and Preah Vihear, where artemisinin resistance is common and dihydroartemisinin-piperaquine has been the frontline treatment for at least 6 years.
“By surveying the piperaquine resistance marker across Southeast Asia in real-time, we can identify those areas where dihydroartemisinin-piperaquine will not be effective, and this could enable national malaria control programs to immediately recommend alternative therapies, such as artesunate-mefloquine,” Dr Fairhurst said.
“This approach will be crucial to eliminating multidrug-resistant parasites in Southeast Asia before they spread to sub-Saharan Africa, where most of the world’s malaria transmission, illness, and death occur.”

Plasmodium falciparum
Image from CDC/Mae Melvin
Two studies have revealed genetic markers associated with resistance to piperaquine, one of the most commonly used malaria drugs in Southeast Asia.
Investigators found that Plasmodium falciparum parasites were less sensitive to piperaquine if they had the exo-E415G variant or multiple copies of the plasmepsin 2 and plasmepsin 3 genes.
Furthermore, the presence of these markers could identify which malaria patients would fail piperaquine treatment.
However, investigators said more research is needed to establish whether plasmepsin gene amplification and the exo-E415G variant actually cause piperaquine resistance and to explore whether this association extends to other parts of the world.
The 2 groups of investigators reported the findings of their studies in The Lancet Infectious Diseases.
About resistance
The combination of artemisinin and piperaquine (dihydroartemisinin-piperaquine) is a frontline treatment for malaria in Southeast Asia.
However, in recent years, the emergence of resistance to these drugs in Cambodia means that up to 60% of patients fail treatment in several provinces. There are fears that resistance could spread to other regions where malaria is endemic.
Genome-wide association studies have been conducted to identify genetic variations among P falciparum parasites that can be linked to variations in drug resistance.
Recent studies have revealed genetic markers linked to resistance to artemisinin (K13) and mefloquine (Pfmdr1), but, until now, markers for resistance to other combination drugs had not been identified.
Plasmepsin genes
In the first study, Didier Ménard, PhD, of the Institut Pasteur in Phnom Penh, Cambodia, and his colleagues found that amplification of plasmepsin genes in the P falciparum genome was linked to piperaquine resistance.
The investigators first analyzed artemisinin-resistant, Cambodian P falciparum parasite lines, comparing the exomes of lines that were piperaquine-susceptible and those that were piperaquine-resistant.
The team said this revealed an increased copy number of the plasmepsin 2-plasmepsin 3 gene cluster as a putative genetic signature associated with piperaquine resistance.
The investigators then confirmed their findings in a set of 725 P falciparum parasites collected from patients across Cambodia since 2009.
Individuals infected with parasites with multiple copies of the plasmepsin 2 gene were, on average, 20.4 times more likely to experience dihydroartemisinin-piperaquine treatment failure.
Furthermore, the geographical distribution and rise in the proportion of parasites with multiple copies of the plasmepsin 2 and 3 genes in Cambodia corresponded with areas that had reported increases in dihydroartemisinin-piperaquine treatment failures in recent years.
“We now know that amplification of the plasmepsin 2 and plasmepsin 3 genes is an important factor in determining how well piperaquine kills malaria parasites,” Dr Ménard said. “A genetic toolkit combining this new marker with markers of artemisinin and mefloquine resistance could be used to track the spread of resistance and provide timely information for containment policies.”
“Piperaquine resistance, although currently confined to Cambodia, is a major concern, because patients suffering malaria are almost untreatable. At present, alternative treatments are scarce, and the reduced cure rates lead to prolonged parasite carriage, increasing the potential of transmitting the resistant parasites and endangering efforts to control and eliminate malaria.”
Exo-E415G variant
The second study was conducted by Rick Fairhurst, MD, of the National Institutes of Health in Bethesda, Maryland, and his colleagues.
The investigators analyzed 486 P falciparum parasites collected between July 9, 2010, and December 31, 2013, from 3 different provinces in Cambodia where artemisinin resistance and dihydroartemisinin-piperaquine treatment failure are common (Pursat), emerging (Preah Vihear), or uncommon (Ratanakiri).
The investigators exposed the parasites to therapeutic levels of piperaquine and measured how well each strain grew. At the same time, they sequenced the entire genome of each parasite.
Parasites were more likely to survive exposure to piperaquine if they had 2 or more copies of the plasmepsin 2 and plasmepsin 3 genes, as well as the exo-E415G variant on chromosome 13.
Similarly, in a further analysis of 133 patient samples, the investigators found that individuals infected with parasites carrying multiple copies of the plasmepsin genes were much more likely to fail treatment with dihydroartemisinin-piperaquine, as were individuals who had parasites with the exo-E415G variant.
The team also noted that the prevalence of exo-E415G and plasmepsin 2-3 markers has risen substantially in recent years in Pursat and Preah Vihear, where artemisinin resistance is common and dihydroartemisinin-piperaquine has been the frontline treatment for at least 6 years.
“By surveying the piperaquine resistance marker across Southeast Asia in real-time, we can identify those areas where dihydroartemisinin-piperaquine will not be effective, and this could enable national malaria control programs to immediately recommend alternative therapies, such as artesunate-mefloquine,” Dr Fairhurst said.
“This approach will be crucial to eliminating multidrug-resistant parasites in Southeast Asia before they spread to sub-Saharan Africa, where most of the world’s malaria transmission, illness, and death occur.”

Plasmodium falciparum
Image from CDC/Mae Melvin
Two studies have revealed genetic markers associated with resistance to piperaquine, one of the most commonly used malaria drugs in Southeast Asia.
Investigators found that Plasmodium falciparum parasites were less sensitive to piperaquine if they had the exo-E415G variant or multiple copies of the plasmepsin 2 and plasmepsin 3 genes.
Furthermore, the presence of these markers could identify which malaria patients would fail piperaquine treatment.
However, investigators said more research is needed to establish whether plasmepsin gene amplification and the exo-E415G variant actually cause piperaquine resistance and to explore whether this association extends to other parts of the world.
The 2 groups of investigators reported the findings of their studies in The Lancet Infectious Diseases.
About resistance
The combination of artemisinin and piperaquine (dihydroartemisinin-piperaquine) is a frontline treatment for malaria in Southeast Asia.
However, in recent years, the emergence of resistance to these drugs in Cambodia means that up to 60% of patients fail treatment in several provinces. There are fears that resistance could spread to other regions where malaria is endemic.
Genome-wide association studies have been conducted to identify genetic variations among P falciparum parasites that can be linked to variations in drug resistance.
Recent studies have revealed genetic markers linked to resistance to artemisinin (K13) and mefloquine (Pfmdr1), but, until now, markers for resistance to other combination drugs had not been identified.
Plasmepsin genes
In the first study, Didier Ménard, PhD, of the Institut Pasteur in Phnom Penh, Cambodia, and his colleagues found that amplification of plasmepsin genes in the P falciparum genome was linked to piperaquine resistance.
The investigators first analyzed artemisinin-resistant, Cambodian P falciparum parasite lines, comparing the exomes of lines that were piperaquine-susceptible and those that were piperaquine-resistant.
The team said this revealed an increased copy number of the plasmepsin 2-plasmepsin 3 gene cluster as a putative genetic signature associated with piperaquine resistance.
The investigators then confirmed their findings in a set of 725 P falciparum parasites collected from patients across Cambodia since 2009.
Individuals infected with parasites with multiple copies of the plasmepsin 2 gene were, on average, 20.4 times more likely to experience dihydroartemisinin-piperaquine treatment failure.
Furthermore, the geographical distribution and rise in the proportion of parasites with multiple copies of the plasmepsin 2 and 3 genes in Cambodia corresponded with areas that had reported increases in dihydroartemisinin-piperaquine treatment failures in recent years.
“We now know that amplification of the plasmepsin 2 and plasmepsin 3 genes is an important factor in determining how well piperaquine kills malaria parasites,” Dr Ménard said. “A genetic toolkit combining this new marker with markers of artemisinin and mefloquine resistance could be used to track the spread of resistance and provide timely information for containment policies.”
“Piperaquine resistance, although currently confined to Cambodia, is a major concern, because patients suffering malaria are almost untreatable. At present, alternative treatments are scarce, and the reduced cure rates lead to prolonged parasite carriage, increasing the potential of transmitting the resistant parasites and endangering efforts to control and eliminate malaria.”
Exo-E415G variant
The second study was conducted by Rick Fairhurst, MD, of the National Institutes of Health in Bethesda, Maryland, and his colleagues.
The investigators analyzed 486 P falciparum parasites collected between July 9, 2010, and December 31, 2013, from 3 different provinces in Cambodia where artemisinin resistance and dihydroartemisinin-piperaquine treatment failure are common (Pursat), emerging (Preah Vihear), or uncommon (Ratanakiri).
The investigators exposed the parasites to therapeutic levels of piperaquine and measured how well each strain grew. At the same time, they sequenced the entire genome of each parasite.
Parasites were more likely to survive exposure to piperaquine if they had 2 or more copies of the plasmepsin 2 and plasmepsin 3 genes, as well as the exo-E415G variant on chromosome 13.
Similarly, in a further analysis of 133 patient samples, the investigators found that individuals infected with parasites carrying multiple copies of the plasmepsin genes were much more likely to fail treatment with dihydroartemisinin-piperaquine, as were individuals who had parasites with the exo-E415G variant.
The team also noted that the prevalence of exo-E415G and plasmepsin 2-3 markers has risen substantially in recent years in Pursat and Preah Vihear, where artemisinin resistance is common and dihydroartemisinin-piperaquine has been the frontline treatment for at least 6 years.
“By surveying the piperaquine resistance marker across Southeast Asia in real-time, we can identify those areas where dihydroartemisinin-piperaquine will not be effective, and this could enable national malaria control programs to immediately recommend alternative therapies, such as artesunate-mefloquine,” Dr Fairhurst said.
“This approach will be crucial to eliminating multidrug-resistant parasites in Southeast Asia before they spread to sub-Saharan Africa, where most of the world’s malaria transmission, illness, and death occur.”
Team targets fructose to treat AML

Photo from the University
of Hawaii Cancer Center
Researchers say they have discovered a novel treatment strategy for acute myeloid leukemia (AML)—inhibiting fructose utilization.
The team discovered that AML cells are prone to using fructose for energy, and increased fructose utilization predicts poor treatment outcomes in AML patients.
The researchers also found that 2,5-anhydro-D-mannitol (2,5-AM) could inhibit fructose use in AML cells and therefore hinder their growth.
Wei Jia, PhD, of the University of Hawaii Cancer Center in Honolulu, and his colleagues reported these findings in Cancer Cell.
The researchers said their findings highlight the unique ability of AML cells to switch their energy supply from glucose to fructose, when glucose is in short supply.
Fructose is the second most abundant blood sugar in the body and is used as a glucose alternative by AML cells to retain energy. After the switch, AML cells begin to multiply faster.
The study revealed a potential treatment by stopping the glucose transporter GLUT5. This restricts the AML energy supply and effectively slows AML growth.
To target GLUT5, the researchers used 2,5-AM, a fructose analog with high affinity for GLUT5.
The team tested 2,5-AM in AML cells with enhanced fructose utilization and found the drug significantly suppressed fructose-induced proliferation, colony growth, and migration in the absence of glucose or when glucose levels were low.
The researchers tested 2,5-AM in 4 different AML cell lines and found the drug suppressed fructose-induced cell proliferation in a dose-dependent manner in all of the cell lines under glucose-limiting conditions. However, 2,5-AM had little effect on glucose-induced cell proliferation.
The team tested 2,5-AM in normal monocytes as well. They said the drug had a negligible effect on glucose-induced cell growth.
“Our normal cells hardly rely on fructose for growth,” Dr Jia noted. “This makes the fructose transport in cancer cells an attractive drug target.”
Finally, the researchers tested 2,5-AM in combination with ara-C in the 4 AML cell lines. They observed a synergistic effect between the drugs in all cell lines in the absence of glucose or when glucose levels were low.
“We are in the process of developing a GLUT5 inhibitor, thus cutting the cancer cells’ energy source and eventually killing them,” Dr Jia said. “The new GLUT5 inhibitor can potentially be used alone or in addition to the current chemotherapy drugs to enhance anticancer effects.”

Photo from the University
of Hawaii Cancer Center
Researchers say they have discovered a novel treatment strategy for acute myeloid leukemia (AML)—inhibiting fructose utilization.
The team discovered that AML cells are prone to using fructose for energy, and increased fructose utilization predicts poor treatment outcomes in AML patients.
The researchers also found that 2,5-anhydro-D-mannitol (2,5-AM) could inhibit fructose use in AML cells and therefore hinder their growth.
Wei Jia, PhD, of the University of Hawaii Cancer Center in Honolulu, and his colleagues reported these findings in Cancer Cell.
The researchers said their findings highlight the unique ability of AML cells to switch their energy supply from glucose to fructose, when glucose is in short supply.
Fructose is the second most abundant blood sugar in the body and is used as a glucose alternative by AML cells to retain energy. After the switch, AML cells begin to multiply faster.
The study revealed a potential treatment by stopping the glucose transporter GLUT5. This restricts the AML energy supply and effectively slows AML growth.
To target GLUT5, the researchers used 2,5-AM, a fructose analog with high affinity for GLUT5.
The team tested 2,5-AM in AML cells with enhanced fructose utilization and found the drug significantly suppressed fructose-induced proliferation, colony growth, and migration in the absence of glucose or when glucose levels were low.
The researchers tested 2,5-AM in 4 different AML cell lines and found the drug suppressed fructose-induced cell proliferation in a dose-dependent manner in all of the cell lines under glucose-limiting conditions. However, 2,5-AM had little effect on glucose-induced cell proliferation.
The team tested 2,5-AM in normal monocytes as well. They said the drug had a negligible effect on glucose-induced cell growth.
“Our normal cells hardly rely on fructose for growth,” Dr Jia noted. “This makes the fructose transport in cancer cells an attractive drug target.”
Finally, the researchers tested 2,5-AM in combination with ara-C in the 4 AML cell lines. They observed a synergistic effect between the drugs in all cell lines in the absence of glucose or when glucose levels were low.
“We are in the process of developing a GLUT5 inhibitor, thus cutting the cancer cells’ energy source and eventually killing them,” Dr Jia said. “The new GLUT5 inhibitor can potentially be used alone or in addition to the current chemotherapy drugs to enhance anticancer effects.”

Photo from the University
of Hawaii Cancer Center
Researchers say they have discovered a novel treatment strategy for acute myeloid leukemia (AML)—inhibiting fructose utilization.
The team discovered that AML cells are prone to using fructose for energy, and increased fructose utilization predicts poor treatment outcomes in AML patients.
The researchers also found that 2,5-anhydro-D-mannitol (2,5-AM) could inhibit fructose use in AML cells and therefore hinder their growth.
Wei Jia, PhD, of the University of Hawaii Cancer Center in Honolulu, and his colleagues reported these findings in Cancer Cell.
The researchers said their findings highlight the unique ability of AML cells to switch their energy supply from glucose to fructose, when glucose is in short supply.
Fructose is the second most abundant blood sugar in the body and is used as a glucose alternative by AML cells to retain energy. After the switch, AML cells begin to multiply faster.
The study revealed a potential treatment by stopping the glucose transporter GLUT5. This restricts the AML energy supply and effectively slows AML growth.
To target GLUT5, the researchers used 2,5-AM, a fructose analog with high affinity for GLUT5.
The team tested 2,5-AM in AML cells with enhanced fructose utilization and found the drug significantly suppressed fructose-induced proliferation, colony growth, and migration in the absence of glucose or when glucose levels were low.
The researchers tested 2,5-AM in 4 different AML cell lines and found the drug suppressed fructose-induced cell proliferation in a dose-dependent manner in all of the cell lines under glucose-limiting conditions. However, 2,5-AM had little effect on glucose-induced cell proliferation.
The team tested 2,5-AM in normal monocytes as well. They said the drug had a negligible effect on glucose-induced cell growth.
“Our normal cells hardly rely on fructose for growth,” Dr Jia noted. “This makes the fructose transport in cancer cells an attractive drug target.”
Finally, the researchers tested 2,5-AM in combination with ara-C in the 4 AML cell lines. They observed a synergistic effect between the drugs in all cell lines in the absence of glucose or when glucose levels were low.
“We are in the process of developing a GLUT5 inhibitor, thus cutting the cancer cells’ energy source and eventually killing them,” Dr Jia said. “The new GLUT5 inhibitor can potentially be used alone or in addition to the current chemotherapy drugs to enhance anticancer effects.”