User login
The use of pharmacogenetic testing to help drive decisions for medication management of patients with psychiatric illnesses is growing. It’s becoming increasingly common for patients or the parents of pediatric patients to request pharmacogenetic testing or to bring the results of prior testing to their appointment. In these situations, patients may ask clinicians to consider the recommendations from these testing reports, which rarely provide guidance specific to pediatric patients. However, this can be difficult for clinicians who did not receive education in pharmacogenetics and may not be familiar with the evidence or options for pharmacogenetic testing. Many of the pharmacogenetic associations identified thus far have been discovered in adults, but studies in pediatric patients are relatively rare. This article reviews pharmacogenetic testing and the evidence supporting it, and describes implementation of routine pharmacogenetics testing at a children’s hospital.
CASE
Testing leads to dose adjustment, improvement
Ms. R, age 16, presents with treatment-resistant major depressive disorder that is characterized by a significant neurovegetative burden and prominent anhedonia, as well as intermittent suicidal ideation without intent or plan. She reportedly did not improve after multiple medication trials, including
Augmentation strategies included
_
Drug metabolism and genetic variants
It is common for patients with psychiatric disorders to receive trials of multiple psychotropic medications prior to identifying one that reduces symptom burden without producing intolerable adverse effects. Due to the high frequency of toxicity-related adverse effects (observed in 20% to 70% of patients),1 these medications are frequently initiated at low doses and titrated slowly until the patient either experiences an intolerable adverse effect or achieves symptomatic remission.1,2 The practice of slow titration at the start of treatment increases the risk of undertreatment in many patients, and may ultimately lead to a medication change due to the lack of response.
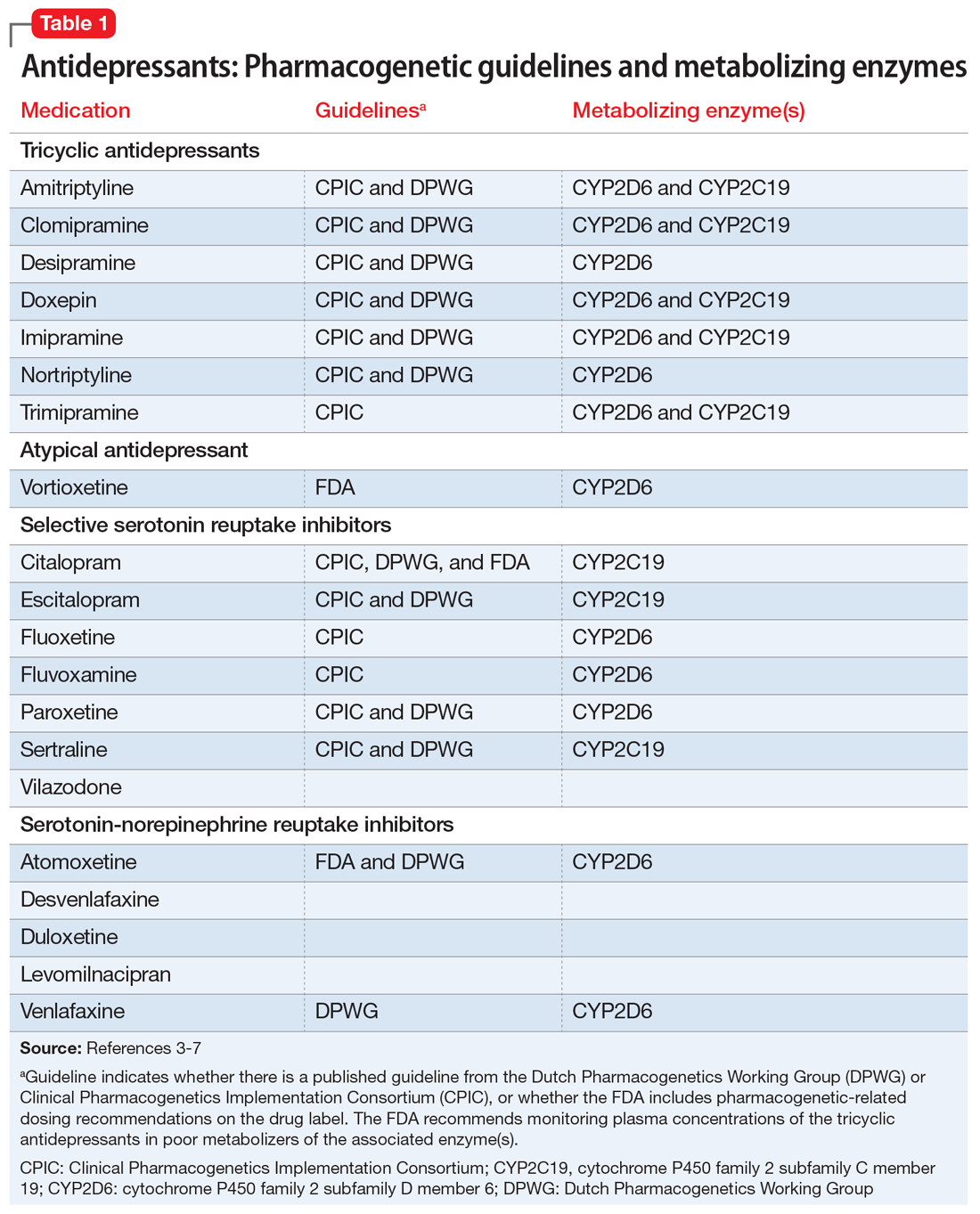
Many of the medications used to treat psychiatric illnesses are primarily metabolized by 2 CYP enzymes expressed in the liver, encoded by the CYP2D6 and CYP2C19 genes3 (Table 13-7 and Table 23,6,7). These drug-metabolizing enzymes affect the pharmacokinetics of many medications. Some medications are converted to an active form by these enzymes, and some are inactivated. The contributions of CYP enzymes to the pharmacokinetics of neuropsychiatric medications have been well-described; however, there is less evidence on whether variants in these genes are associated with treatment efficacy, especially in pediatric patients.8,9 CYP2D6 enzyme activity reaches adult levels soon after birth, but children may have higher CYP2C19 activity than adults.4 CYP3A4 also contributes to the metabolism of many medications; however, there is only weak evidence that genetic variants in CYP3A4 contribute to variability in the pharmacokinetics of these medications, and there are currently no dosing guidelines based on pharmacogenetics available for this gene.10
As is common in the pharmacogenetic field, genotypes are denoted with a “star allele” (eg, *2) rather than positional nomenclature (eg, c.681G>A). The normal allele is usually designated as *1, and this result is given in the absence of the tested alleles. There is no consensus on the minimum set of alleles to be tested for most genes,11 so commercially available tests vary widely in what alleles are tested (and therefore what they exclude before calling a normal allele).12 The metabolizer phenotype for a patient is determined by taking into account the activity of each of the patient’s 2 alleles (eg, *1/*2). A patient is categorized as a poor-, intermediate-, normal- (extensive-), or ultra-rapid metabolizer. Generally, the allele definitions are widely agreed upon (what genetic variant or variants comprise the *2 allele) due to nomenclature committees for each gene; however, because there are no standards for interpretation, the interpretation of the activity of the alleles and conversion to metabolizer phenotype varies among clinics.13
Continue to: Guidelines help with genotype-guided dosing
Guidelines help with genotype-guided dosing

Each CPIC guideline specifically addresses use in pediatric patients, indicating that there are relatively few studies in pediatrics, but “it may be appropriate to extrapolate these recommendations to adolescents or possibly younger children with close monitoring.”4 The DPWG guidelines do not mention whether or not the recommendations are applicable to children. Neither CPIC nor the DPWG provides guidance on when to test; however, the French National Network of Pharmacogenetics (Réseau national de pharmacogénétique) recommends CYP2D6 and CYP2C19 genotyping before initiating antidepressant treatment, especially in patients with a high risk of toxicity.16
In the case above, Ms. R was determined to be a CYP2D6 ultra-rapid metabolizer. Because she showed some initial response to aripiprazole and venlafaxine ER, which are both metabolized by CYP2D6, these medications were very quickly titrated up, and the increased dosages produced the desired response. Venlafaxine is metabolized to the active metabolite O-desmethylvenlafaxine by CYP2D6. The DPWG recommends increasing the dose of venlafaxine in CYP2D6 ultra-rapid metabolizers to 150% of the normal dose based on the decreased serum concentrations of venlafaxine and O-desmethylvenlafaxine in these patients.6 Aripiprazole is also metabolized by CYP2D6; however, the FDA and DPWG give no recommendations for ultra-rapid metabolizers, but do recommend reducing the dose of aripiprazole in CYP2D6 poor metabolizers.
Multiple studies in adults have analyzed the association between pharmacokinetic (CYP2D6 and CYP2C19) or pharmacodynamic genes (SLC6A4, HTR2A, and GRIK4) and outcomes,17 including some large clinical trials that conducted genome-wide association studies18-20 and meta-analyses across multiple studies.21,22 Most pharmacogenetic studies in psychiatric patients are small, and very few have included pediatric patients. However, with more interest in neuropsychiatric pharmacogenetics, these studies are becoming more common.23-26
Continue to: Limited evidence from studies of commercially available tests
Limited evidence from studies of commercially available tests
Several pharmacogenetic tests are commercially available, including some that focus on providing information that can be used specifically when prescribing psychiatric medications, such as the GeneSight Psychotropic test, CNSdose, Genomind, and Neuropharmagen.
In an industry-sponsored, nonrandomized clinical trial that included patients for whom prescribing decisions were made based on the GeneSight test, outcomes in adults were improved compared with treatment as usual,27 inpatient stays were shorter,28 and pharmacy costs were reduced.29 In one of these studies, the authors noted that the traditional, single-gene analysis was not associated with improved outcomes, whereas the multiple gene combination (pharmacokinetic and pharmacodynamic genes) was associated with improved outcomes among patients with depression.27 However, when GeneSightwas compared with treatment as usual in a small randomized trial, there was not a significant association between use of the test and improved outcomes among patients with treatment-resistant depression.30 The results of a much larger randomized trial (N = 1,167) are available31 and expected to be published, but patients younger than age 18 were excluded from this study.32 A retrospective study conducted in adult psychiatric patients found that patients whose treatment followed recommendations of a pharmacogenetic test including 20 genes were almost 4 times more likely to improve than patients whose treatment did not follow the recommendations.33
Pharmacogenetic testing at our pediatric inpatient unit
The Cincinnati Children’s Division of Child and Adolescent Psychiatry is the largest psychiatric inpatient service in a U.S. pediatric hospital. Starting in 2004, we adopted pharmacogenetically-guided dosing of psychiatric medications.34 CYP2D6 and CYP2C19 were chosen for testing because the enzymes encoded by these genes metabolize many of the antidepressants and antipsychotics that patients admitted to our unit will receive, and the clinicians wanted all available tools to help improve the care of these patients. To date, the Genetic Pharmacology Service (GPS) has performed >25,000 tests for variants in CYP2D6 and CYP2C19 as part of inpatient care. Patients provide a specimen (blood or buccal swab) at the time of admission to inpatient psychiatry, genotyping is performed onsite by the Molecular Genetics Laboratory (certified by the College of American Pathologists [CAP]/Clinical Laboratory Improvement Amendments [CLIA]) and the results are posted to the medical record within 2 business days. The report contains the patient’s alleles for CYP2D6 and CYP2C19, the genotype-predicted metabolizer phenotype, and dosing recommendations for 19 drugs (provided as a percentage of the standard dose). Insurance is billed for the test, and reimbursement is usually received when the test is performed as part of an inpatient stay.
The GPS team performed a retrospective chart review after the first panel was implemented in 2005.23 The study included 279 patients who were receiving a medication metabolized by one of the 2 genes tested. The poor metabolizers had the highest efficacy and highest number of adverse drug reactions, while ultra-rapid metabolizers had the lowest efficacy and lowest number of adverse reactions during their initial inpatient stay. In patients not treated with medications metabolized by CYP2D6 or CYP2C19, there was no association between metabolizer status and efficacy or adverse drug reactions. In this retrospective study, there was no association between metabolizer status and length of stay.
Overcoming the challenges
One challenge with many of the pharmacogenetic tests is interpretation of the results. The reports can span more than 20 pages, and clinicians may not have time to thoroughly read and understand how best to use all of this information. Sometimes the reports can make it seem like the first-line medication for the patient’s condition is not the best choice, but it could work well when dosed appropriately based on the patient’s genotype. Each commercially available test has a different way of presenting results,13 so when choosing a pharmacogenetic test, one should be sure to see a sample report. Vo et al35 recently reviewed factors to consider when choosing a pharmacogenetic test.
Continue to: Because patients and families also have difficulty understanding the reports...
Because patients and families also have difficulty understanding the reports, we created patient education sheets,36 written at an eighth grade level with feedback from parents and modeled on those provided by St. Jude Children’s Research Hospital.37 St. Jude Children’s Research Hospital also has pharmacogenetic competencies that pharmacists and nurses must pass.38,39 The following is a sample explanation that one of our nurses uses to educate parents on what is being tested and what effect the results will have on the treatment plan.
“During your child’s stay we will be completing a genetic test to help us understand how he/she processes the types of medications that we may be likely to start during their hospitalization. This does not tell us which medication will be best—unfortunately within the field of psychiatry there is still some unavoidable trial and error; rather, what it will do is tell us how to make sure that the dosing is at a level that would be safe for the way your child’s body breaks down the medicine, so that he/she can get the intended benefit of the medicine’s effects, while decreasing the risk of uncomfortable side effects, where possible.”
Other challenges in pharmacogenetic testing are the cost, disease risk, and concern about how genetic information will be used. Because these tests are often not covered by health insurance, some commercial pharmacogenetic testing companies offer an out-of-pocket maximum in the $250 to $350 range to reduce the cost to the patient. Some pharmacogenetic testing companies also test for genes associated with disease, so if a clinician orders the test, he or she may be responsible for sharing that information with the patient. For most pharmacogenetic testing companies, the turn-around time is 2 to 10 days. Genetic information is protected by federal laws, including Genetic Information Nondiscrimination Act (GINA) and Health Insurance Portability and Accountability Act (HIPAA).
The choice of psychotropic medication is complex, and although we would like pharmacogenetics to be the only answer to why every patient does or does not respond to a medication, it is not. Response to medication is influenced by age, comorbidities, illness severity, illness duration, compliance, gender, concomitant medications, and potentially more.40 Pharmacogenetics is another tool at the clinician’s disposal to help in choosing a medication and dose. There is a clear association between CYP2D6 and CYP2C19 and exposure to many antidepressants and antipsychotics (reviewed by Stingl et al3); however, the link between exposure and response is much weaker. It may be strengthened by the inclusion of pharmacodynamic information (the level of expression of the drug target), which can be influenced by genetic variants.41 At the present time, the most evidence exists for testing CYP2D6 and CYP2C19, and the CPIC4,5,15 and DWPG6 guidelines provide evidence-based recommendations for how to adjust medication dosages based on the results.
There is clearly much more research that needs to be done in the field of neuropsychiatric pharmacogenetics, especially in pediatric populations. As we see increased utilization of pharmacogenetic tests in psychiatry, there is also a need for pharmacogenetic education of patients, families, nurses, pharmacists, and psychiatrists. Several good pharmacogenetic resources that contain up-to-date summaries of the available evidence linking pharmacogenetic variants to medication response, implementation resources, and educational resources are available. These include CPIC (www.cpicpgx.org), PharmGKB (www.pharmgkb.org), and the IGNITE Spark Toolbox (https://ignite-genomics.org/spark-toolbox/clinicians/).
Acknowledgements
The author thanks Jen Milau, APRN, for the case study and sample explanation, and Jeffrey Strawn, MD, FAACP, Ethan Poweleit, and Stacey Aldrich, MS, for help with preparing this manuscript.
1. Cipriani A, Zhou X, Del Giovane C, et al. Comparative efficacy and tolerability of antidepressants for major depressive disorder in children and adolescents: a network meta-analysis. Lancet. 2016; 388(10047):881-890.
2. Correll CU, Sheridan EM, DelBello MP. Antipsychotic and mood stabilizer efficacy and tolerability in pediatric and adult patients with bipolar I mania: a comparative analysis of acute, randomized, placebo-controlled trials. Bipolar Disord. 2010;12(2):116-141.
3. Stingl JC, Brockmoller J, Viviani R. Genetic variability of drug-metabolizing enzymes: the dual impact on psychiatric therapy and regulation of brain function. Mol Psychiatry. 2013;18(3):273-287.
4. Hicks JK, Bishop JR, Sangkuhl K, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin Pharmacol Ther. 2015;98(2):127-134.
5. Hicks JK, Sangkuhl K, Swen JJ, et al. Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin Pharmacol Ther. 2017;102(1):37-44.
6. Swen JJ, Nijenhuis M, de Boer A, et al. Pharmacogenetics: from bench to byte--an update of guidelines. Clin Pharmacol Ther. 2011;89(5):662-673.
7. Swen JJ, Wilting I, de Goede AL, et al. Pharmacogenetics: from bench to byte. Clin Pharmacol Ther. 2008;83(5):781-787.
8. GENDEP Investigators, MARS Investigators, and STAR*D Investigators. Common genetic variation and antidepressant efficacy in major depressive disorder: a meta-analysis of three genome-wide pharmacogenetic studies. Am J Psychiatry. 2013;170(2):207-217.
9. Ji Y, Schaid DJ, Desta Z, et al. Citalopram and escitalopram plasma drug and metabolite concentrations: genome-wide associations. Br J Clin Pharmacol. 2014;78(2):373-383.
10. Werk AN, Cascorbi I. Functionalgene variants of CYP3A4. Clin Pharmacol Ther. 2014:96(3):340-348.
11. Pratt VM, Del Tredici AL, Hachad H, et al. Recommendations for clinical CYP2C19 genotyping allele selection: a report of the Association for Molecular Pathology. J Mol Diagn. 2018;20(3):269-276.
12. Bousman CA, Jaksa P, Pantelis C. Systematic evaluation of commercial pharmacogenetic testing in psychiatry: a focus on CYP2D6 and CYP2C19 allele coverage and results reporting. Pharmacogenet Genomics. 2017;27(11):387-393.
13. Hicks JK, Swen JJ, Gaedigk A. Challenges in CYP2D6 phenotype assignment from genotype data: a critical assessment and call for standardization. Curr Drug Metab. 2014;15(2):218-232.
14. Caudle KE, Klein TE, Hoffman JM, et al. Incorporation of pharmacogenomics into routine clinical practice: the Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline development process. Curr Drug Metab. 2014;15(2):209-217.
15. Hicks JK, Swen JJ, Thorn CF, et al. Clinical Pharmacogenetics Implementation Consortium guideline for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants. Clin Pharmacol Ther. 2013;93(5):402-408.
16. Quaranta S, Dupouey J, Colle R, et al. Pharmacogenetics of antidepressant drugs: State of the art and clinical implementation - recommendations from the French National Network of Pharmacogenetics. Therapie. 2017;72(2):311-318.
17. Fabbri C, Minarini A, Nitsu T, et al. Understanding the pharmacogenetics of selective serotonin reuptake inhibitors. Expert Opin Drug Metab Toxicol. 2014;10(8):1093-1118.
18. Mrazek DA, Rush AJ, Biernacka JM, et al. SLC6A4 variation and citalopram response. Am J Med Genet B Neuropsychiatr Genet. 2009;150B(3):341-351.
19. Biernacka JM, Sangkuhl K, Jenkins G, et al. The International SSRI Pharmacogenomics Consortium (ISPC): a genome-wide association study of antidepressant treatment response. Transl Psychiatry. 2015;5:e553. doi: 10.1038/tp.2015.47.
20. Horstmann S, Lucae S, Menke A, et al. Polymorphisms in GRIK4, HTR2A, and FKBP5 show interactive effects in predicting remission to antidepressant treatment. Neuropsychopharmacology. 2010;35(3):727-740.
21. Porcelli S, Fabbri C, Serretti A. Meta-analysis of serotonin transporter gene promoter polymorphism (5-HTTLPR) association with antidepressant efficacy. Eur Neuropsychopharmacol. 2012;22(4):239-258.
22. Niitsu T, Fabbri C, Bentini F, et al. Pharmacogenetics in major depression: a comprehensive meta-analysis. Prog Neuropsychopharmacol Biol Psychiatry. 2013;45:183-194.
23. Prows CA, Nick TG, Saldaña SN, et al. Drug-metabolizing enzyme genotypes and aggressive behavior treatment response in hospitalized pediatric psychiatric patients. J Child Adolesc Psychopharmacol. 2009;19(4):385-394.
24. Rotberg B, Kronenberg S, Carmel M, et al. Additive effects of 5-HTTLPR (serotonin transporter) and tryptophan hydroxylase 2 G-703T gene polymorphisms on the clinical response to citalopram among children and adolescents with depression and anxiety disorders. J Child Adolesc Psychopharmacol. 2013;23(2):117-122.
25. Kronenberg S, Apter A, Brent D, et al. Serotonin transporter polymorphism (5-HTTLPR) and citalopram effectiveness and side effects in children with depression and/or anxiety disorders. J Child Adolesc Psychopharmacol. 2007;17(6):741-750.
26. AlOlaby RR, Sweha SR, Silva M, et al. Molecular biomarkers predictive of sertraline treatment response in young children with fragile X syndrome. Brain Dev. 2017;39(6):483-492.
27. Altar CA, Carhart JM, Allen JD, et al. Clinical validity: Combinatorial pharmacogenomics predicts antidepressant responses and healthcare utilizations better than single gene phenotypes. Pharmacogenomics J. 2015;15(5):443-451.
28. Winner J, Allen JD, Altar CA, et al. Psychiatric pharmacogenomics predicts health resource utilization of outpatients with anxiety and depression. Transl Psychiatry. 2013;3:e242. doi:10.1038/tp.2013.2.
29. Winner JG, Carhart JM, Altar CA, et al. Combinatorial pharmacogenomic guidance for psychiatric medications reduces overall pharmacy costs in a 1 year prospective evaluation. Curr Med Res Opin. 2015;31(9):1633-1643.
30. Winner JG, Carhart JM, Altar CA, et al. A prospective, randomized, double-blind study assessing the clinical impact of integrated pharmacogenomic testing for major depressive disorder. Discov Med. 2013;16(89):219-227.
31. Genesight. GUIDED clinical study. https://genesight.com/greden-study/. Updated May 31, 2018. Accessed August 1, 2018.
32. U.S. National Library of Medicine ClinicalTrials.gov. Genomics used to improve DEpression decisions (GUIDED). https://clinicaltrials.gov/ct2/show/NCT02109939. Accessed July 24, 2018.
33. Espadaler J, Tuson M, Lopez-Ibor JM, et al. Pharmacogenetic testing for the guidance of psychiatric treatment: a multicenter retrospective analysis. CNS Spectrums. 2017;22(4):315-324.
34. Ramsey LB, Prows CA, Zhang K, et al. Implementation of pharmacogenetics at Cincinnati Children’s Hospital Medical Center: lessons learned over 14 years of personalizing medicine. Clin Pharmacol Ther. 2018. doi: 10.1002/cpt.1165. [Epub ahead of print].
35. Vo TT, Bell GC, Owusu Obeng A, et al. Pharmacogenomics implementation: considerations for selecting a reference laboratory. Pharmacotherapy. 2017;37(9):1014-1022.
36. Cincinnati Children’s Hospital. Genetic Pharmacology Service: Education. www.cincinnatichildrens.org/gpsinfo. Accessed August 1, 2018.
37. St. Jude Children’s Research Hospital. Do You Know...Cytochrome P450 2D6 (CYP2D6) and medicines. https://www.stjude.org/treatment/patient-resources/caregiver-resources/patient-family-education-sheets/pharmacy-and-medicines/cytochrome-p450-2d6-cyp2d6-and-medicines.html. Accessed August 1, 2018.
38. St. Jude Children’s Research Hospital. Implementation Resources for Professionals: Clinical Pharmacogenetics at St. Jude. https://www.stjude.org/research/clinical-trials/pg4kds-pharmaceutical-science/implementation-resources-for-professionals.html. Accessed August 1, 2018.
39. Hoffman JM, Haider CE, Wilkinson MR, et al. PG4KDS: a model for the clinical implementation of pre-emptive pharmacogenetics. Am J Med Genet C Semin Med Genet. 2014;166C(1):45-55.
40. Wehry AM, Ramsey LB, Dulemba SE, et al. Pharmacogenomic testing in child and adolescent psychiatry: an evidence-based review. Curr Probl Pediatr Adolesc Health Care. 2018;48(2):40-49.
41. Tomita T, Yasui-Furukori N, Nakagami T, et al. The influence of 5-HTTLPR genotype on the association between the plasma concentration and therapeutic effect of paroxetine in patients with major depressive disorder. PLoS One. 2014;9(5):e98099. doi: 10.1371/journal.pone.0098099.
The use of pharmacogenetic testing to help drive decisions for medication management of patients with psychiatric illnesses is growing. It’s becoming increasingly common for patients or the parents of pediatric patients to request pharmacogenetic testing or to bring the results of prior testing to their appointment. In these situations, patients may ask clinicians to consider the recommendations from these testing reports, which rarely provide guidance specific to pediatric patients. However, this can be difficult for clinicians who did not receive education in pharmacogenetics and may not be familiar with the evidence or options for pharmacogenetic testing. Many of the pharmacogenetic associations identified thus far have been discovered in adults, but studies in pediatric patients are relatively rare. This article reviews pharmacogenetic testing and the evidence supporting it, and describes implementation of routine pharmacogenetics testing at a children’s hospital.
CASE
Testing leads to dose adjustment, improvement
Ms. R, age 16, presents with treatment-resistant major depressive disorder that is characterized by a significant neurovegetative burden and prominent anhedonia, as well as intermittent suicidal ideation without intent or plan. She reportedly did not improve after multiple medication trials, including
Augmentation strategies included
_
Drug metabolism and genetic variants
It is common for patients with psychiatric disorders to receive trials of multiple psychotropic medications prior to identifying one that reduces symptom burden without producing intolerable adverse effects. Due to the high frequency of toxicity-related adverse effects (observed in 20% to 70% of patients),1 these medications are frequently initiated at low doses and titrated slowly until the patient either experiences an intolerable adverse effect or achieves symptomatic remission.1,2 The practice of slow titration at the start of treatment increases the risk of undertreatment in many patients, and may ultimately lead to a medication change due to the lack of response.
Many of the medications used to treat psychiatric illnesses are primarily metabolized by 2 CYP enzymes expressed in the liver, encoded by the CYP2D6 and CYP2C19 genes3 (Table 13-7 and Table 23,6,7). These drug-metabolizing enzymes affect the pharmacokinetics of many medications. Some medications are converted to an active form by these enzymes, and some are inactivated. The contributions of CYP enzymes to the pharmacokinetics of neuropsychiatric medications have been well-described; however, there is less evidence on whether variants in these genes are associated with treatment efficacy, especially in pediatric patients.8,9 CYP2D6 enzyme activity reaches adult levels soon after birth, but children may have higher CYP2C19 activity than adults.4 CYP3A4 also contributes to the metabolism of many medications; however, there is only weak evidence that genetic variants in CYP3A4 contribute to variability in the pharmacokinetics of these medications, and there are currently no dosing guidelines based on pharmacogenetics available for this gene.10

As is common in the pharmacogenetic field, genotypes are denoted with a “star allele” (eg, *2) rather than positional nomenclature (eg, c.681G>A). The normal allele is usually designated as *1, and this result is given in the absence of the tested alleles. There is no consensus on the minimum set of alleles to be tested for most genes,11 so commercially available tests vary widely in what alleles are tested (and therefore what they exclude before calling a normal allele).12 The metabolizer phenotype for a patient is determined by taking into account the activity of each of the patient’s 2 alleles (eg, *1/*2). A patient is categorized as a poor-, intermediate-, normal- (extensive-), or ultra-rapid metabolizer. Generally, the allele definitions are widely agreed upon (what genetic variant or variants comprise the *2 allele) due to nomenclature committees for each gene; however, because there are no standards for interpretation, the interpretation of the activity of the alleles and conversion to metabolizer phenotype varies among clinics.13
Continue to: Guidelines help with genotype-guided dosing
Guidelines help with genotype-guided dosing

Each CPIC guideline specifically addresses use in pediatric patients, indicating that there are relatively few studies in pediatrics, but “it may be appropriate to extrapolate these recommendations to adolescents or possibly younger children with close monitoring.”4 The DPWG guidelines do not mention whether or not the recommendations are applicable to children. Neither CPIC nor the DPWG provides guidance on when to test; however, the French National Network of Pharmacogenetics (Réseau national de pharmacogénétique) recommends CYP2D6 and CYP2C19 genotyping before initiating antidepressant treatment, especially in patients with a high risk of toxicity.16
In the case above, Ms. R was determined to be a CYP2D6 ultra-rapid metabolizer. Because she showed some initial response to aripiprazole and venlafaxine ER, which are both metabolized by CYP2D6, these medications were very quickly titrated up, and the increased dosages produced the desired response. Venlafaxine is metabolized to the active metabolite O-desmethylvenlafaxine by CYP2D6. The DPWG recommends increasing the dose of venlafaxine in CYP2D6 ultra-rapid metabolizers to 150% of the normal dose based on the decreased serum concentrations of venlafaxine and O-desmethylvenlafaxine in these patients.6 Aripiprazole is also metabolized by CYP2D6; however, the FDA and DPWG give no recommendations for ultra-rapid metabolizers, but do recommend reducing the dose of aripiprazole in CYP2D6 poor metabolizers.
Multiple studies in adults have analyzed the association between pharmacokinetic (CYP2D6 and CYP2C19) or pharmacodynamic genes (SLC6A4, HTR2A, and GRIK4) and outcomes,17 including some large clinical trials that conducted genome-wide association studies18-20 and meta-analyses across multiple studies.21,22 Most pharmacogenetic studies in psychiatric patients are small, and very few have included pediatric patients. However, with more interest in neuropsychiatric pharmacogenetics, these studies are becoming more common.23-26
Continue to: Limited evidence from studies of commercially available tests
Limited evidence from studies of commercially available tests
Several pharmacogenetic tests are commercially available, including some that focus on providing information that can be used specifically when prescribing psychiatric medications, such as the GeneSight Psychotropic test, CNSdose, Genomind, and Neuropharmagen.
In an industry-sponsored, nonrandomized clinical trial that included patients for whom prescribing decisions were made based on the GeneSight test, outcomes in adults were improved compared with treatment as usual,27 inpatient stays were shorter,28 and pharmacy costs were reduced.29 In one of these studies, the authors noted that the traditional, single-gene analysis was not associated with improved outcomes, whereas the multiple gene combination (pharmacokinetic and pharmacodynamic genes) was associated with improved outcomes among patients with depression.27 However, when GeneSightwas compared with treatment as usual in a small randomized trial, there was not a significant association between use of the test and improved outcomes among patients with treatment-resistant depression.30 The results of a much larger randomized trial (N = 1,167) are available31 and expected to be published, but patients younger than age 18 were excluded from this study.32 A retrospective study conducted in adult psychiatric patients found that patients whose treatment followed recommendations of a pharmacogenetic test including 20 genes were almost 4 times more likely to improve than patients whose treatment did not follow the recommendations.33
Pharmacogenetic testing at our pediatric inpatient unit
The Cincinnati Children’s Division of Child and Adolescent Psychiatry is the largest psychiatric inpatient service in a U.S. pediatric hospital. Starting in 2004, we adopted pharmacogenetically-guided dosing of psychiatric medications.34 CYP2D6 and CYP2C19 were chosen for testing because the enzymes encoded by these genes metabolize many of the antidepressants and antipsychotics that patients admitted to our unit will receive, and the clinicians wanted all available tools to help improve the care of these patients. To date, the Genetic Pharmacology Service (GPS) has performed >25,000 tests for variants in CYP2D6 and CYP2C19 as part of inpatient care. Patients provide a specimen (blood or buccal swab) at the time of admission to inpatient psychiatry, genotyping is performed onsite by the Molecular Genetics Laboratory (certified by the College of American Pathologists [CAP]/Clinical Laboratory Improvement Amendments [CLIA]) and the results are posted to the medical record within 2 business days. The report contains the patient’s alleles for CYP2D6 and CYP2C19, the genotype-predicted metabolizer phenotype, and dosing recommendations for 19 drugs (provided as a percentage of the standard dose). Insurance is billed for the test, and reimbursement is usually received when the test is performed as part of an inpatient stay.
The GPS team performed a retrospective chart review after the first panel was implemented in 2005.23 The study included 279 patients who were receiving a medication metabolized by one of the 2 genes tested. The poor metabolizers had the highest efficacy and highest number of adverse drug reactions, while ultra-rapid metabolizers had the lowest efficacy and lowest number of adverse reactions during their initial inpatient stay. In patients not treated with medications metabolized by CYP2D6 or CYP2C19, there was no association between metabolizer status and efficacy or adverse drug reactions. In this retrospective study, there was no association between metabolizer status and length of stay.
Overcoming the challenges
One challenge with many of the pharmacogenetic tests is interpretation of the results. The reports can span more than 20 pages, and clinicians may not have time to thoroughly read and understand how best to use all of this information. Sometimes the reports can make it seem like the first-line medication for the patient’s condition is not the best choice, but it could work well when dosed appropriately based on the patient’s genotype. Each commercially available test has a different way of presenting results,13 so when choosing a pharmacogenetic test, one should be sure to see a sample report. Vo et al35 recently reviewed factors to consider when choosing a pharmacogenetic test.
Continue to: Because patients and families also have difficulty understanding the reports...
Because patients and families also have difficulty understanding the reports, we created patient education sheets,36 written at an eighth grade level with feedback from parents and modeled on those provided by St. Jude Children’s Research Hospital.37 St. Jude Children’s Research Hospital also has pharmacogenetic competencies that pharmacists and nurses must pass.38,39 The following is a sample explanation that one of our nurses uses to educate parents on what is being tested and what effect the results will have on the treatment plan.
“During your child’s stay we will be completing a genetic test to help us understand how he/she processes the types of medications that we may be likely to start during their hospitalization. This does not tell us which medication will be best—unfortunately within the field of psychiatry there is still some unavoidable trial and error; rather, what it will do is tell us how to make sure that the dosing is at a level that would be safe for the way your child’s body breaks down the medicine, so that he/she can get the intended benefit of the medicine’s effects, while decreasing the risk of uncomfortable side effects, where possible.”
Other challenges in pharmacogenetic testing are the cost, disease risk, and concern about how genetic information will be used. Because these tests are often not covered by health insurance, some commercial pharmacogenetic testing companies offer an out-of-pocket maximum in the $250 to $350 range to reduce the cost to the patient. Some pharmacogenetic testing companies also test for genes associated with disease, so if a clinician orders the test, he or she may be responsible for sharing that information with the patient. For most pharmacogenetic testing companies, the turn-around time is 2 to 10 days. Genetic information is protected by federal laws, including Genetic Information Nondiscrimination Act (GINA) and Health Insurance Portability and Accountability Act (HIPAA).
The choice of psychotropic medication is complex, and although we would like pharmacogenetics to be the only answer to why every patient does or does not respond to a medication, it is not. Response to medication is influenced by age, comorbidities, illness severity, illness duration, compliance, gender, concomitant medications, and potentially more.40 Pharmacogenetics is another tool at the clinician’s disposal to help in choosing a medication and dose. There is a clear association between CYP2D6 and CYP2C19 and exposure to many antidepressants and antipsychotics (reviewed by Stingl et al3); however, the link between exposure and response is much weaker. It may be strengthened by the inclusion of pharmacodynamic information (the level of expression of the drug target), which can be influenced by genetic variants.41 At the present time, the most evidence exists for testing CYP2D6 and CYP2C19, and the CPIC4,5,15 and DWPG6 guidelines provide evidence-based recommendations for how to adjust medication dosages based on the results.
There is clearly much more research that needs to be done in the field of neuropsychiatric pharmacogenetics, especially in pediatric populations. As we see increased utilization of pharmacogenetic tests in psychiatry, there is also a need for pharmacogenetic education of patients, families, nurses, pharmacists, and psychiatrists. Several good pharmacogenetic resources that contain up-to-date summaries of the available evidence linking pharmacogenetic variants to medication response, implementation resources, and educational resources are available. These include CPIC (www.cpicpgx.org), PharmGKB (www.pharmgkb.org), and the IGNITE Spark Toolbox (https://ignite-genomics.org/spark-toolbox/clinicians/).
Acknowledgements
The author thanks Jen Milau, APRN, for the case study and sample explanation, and Jeffrey Strawn, MD, FAACP, Ethan Poweleit, and Stacey Aldrich, MS, for help with preparing this manuscript.
The use of pharmacogenetic testing to help drive decisions for medication management of patients with psychiatric illnesses is growing. It’s becoming increasingly common for patients or the parents of pediatric patients to request pharmacogenetic testing or to bring the results of prior testing to their appointment. In these situations, patients may ask clinicians to consider the recommendations from these testing reports, which rarely provide guidance specific to pediatric patients. However, this can be difficult for clinicians who did not receive education in pharmacogenetics and may not be familiar with the evidence or options for pharmacogenetic testing. Many of the pharmacogenetic associations identified thus far have been discovered in adults, but studies in pediatric patients are relatively rare. This article reviews pharmacogenetic testing and the evidence supporting it, and describes implementation of routine pharmacogenetics testing at a children’s hospital.
CASE
Testing leads to dose adjustment, improvement
Ms. R, age 16, presents with treatment-resistant major depressive disorder that is characterized by a significant neurovegetative burden and prominent anhedonia, as well as intermittent suicidal ideation without intent or plan. She reportedly did not improve after multiple medication trials, including
Augmentation strategies included
_
Drug metabolism and genetic variants
It is common for patients with psychiatric disorders to receive trials of multiple psychotropic medications prior to identifying one that reduces symptom burden without producing intolerable adverse effects. Due to the high frequency of toxicity-related adverse effects (observed in 20% to 70% of patients),1 these medications are frequently initiated at low doses and titrated slowly until the patient either experiences an intolerable adverse effect or achieves symptomatic remission.1,2 The practice of slow titration at the start of treatment increases the risk of undertreatment in many patients, and may ultimately lead to a medication change due to the lack of response.
Many of the medications used to treat psychiatric illnesses are primarily metabolized by 2 CYP enzymes expressed in the liver, encoded by the CYP2D6 and CYP2C19 genes3 (Table 13-7 and Table 23,6,7). These drug-metabolizing enzymes affect the pharmacokinetics of many medications. Some medications are converted to an active form by these enzymes, and some are inactivated. The contributions of CYP enzymes to the pharmacokinetics of neuropsychiatric medications have been well-described; however, there is less evidence on whether variants in these genes are associated with treatment efficacy, especially in pediatric patients.8,9 CYP2D6 enzyme activity reaches adult levels soon after birth, but children may have higher CYP2C19 activity than adults.4 CYP3A4 also contributes to the metabolism of many medications; however, there is only weak evidence that genetic variants in CYP3A4 contribute to variability in the pharmacokinetics of these medications, and there are currently no dosing guidelines based on pharmacogenetics available for this gene.10

As is common in the pharmacogenetic field, genotypes are denoted with a “star allele” (eg, *2) rather than positional nomenclature (eg, c.681G>A). The normal allele is usually designated as *1, and this result is given in the absence of the tested alleles. There is no consensus on the minimum set of alleles to be tested for most genes,11 so commercially available tests vary widely in what alleles are tested (and therefore what they exclude before calling a normal allele).12 The metabolizer phenotype for a patient is determined by taking into account the activity of each of the patient’s 2 alleles (eg, *1/*2). A patient is categorized as a poor-, intermediate-, normal- (extensive-), or ultra-rapid metabolizer. Generally, the allele definitions are widely agreed upon (what genetic variant or variants comprise the *2 allele) due to nomenclature committees for each gene; however, because there are no standards for interpretation, the interpretation of the activity of the alleles and conversion to metabolizer phenotype varies among clinics.13
Continue to: Guidelines help with genotype-guided dosing
Guidelines help with genotype-guided dosing

Each CPIC guideline specifically addresses use in pediatric patients, indicating that there are relatively few studies in pediatrics, but “it may be appropriate to extrapolate these recommendations to adolescents or possibly younger children with close monitoring.”4 The DPWG guidelines do not mention whether or not the recommendations are applicable to children. Neither CPIC nor the DPWG provides guidance on when to test; however, the French National Network of Pharmacogenetics (Réseau national de pharmacogénétique) recommends CYP2D6 and CYP2C19 genotyping before initiating antidepressant treatment, especially in patients with a high risk of toxicity.16
In the case above, Ms. R was determined to be a CYP2D6 ultra-rapid metabolizer. Because she showed some initial response to aripiprazole and venlafaxine ER, which are both metabolized by CYP2D6, these medications were very quickly titrated up, and the increased dosages produced the desired response. Venlafaxine is metabolized to the active metabolite O-desmethylvenlafaxine by CYP2D6. The DPWG recommends increasing the dose of venlafaxine in CYP2D6 ultra-rapid metabolizers to 150% of the normal dose based on the decreased serum concentrations of venlafaxine and O-desmethylvenlafaxine in these patients.6 Aripiprazole is also metabolized by CYP2D6; however, the FDA and DPWG give no recommendations for ultra-rapid metabolizers, but do recommend reducing the dose of aripiprazole in CYP2D6 poor metabolizers.
Multiple studies in adults have analyzed the association between pharmacokinetic (CYP2D6 and CYP2C19) or pharmacodynamic genes (SLC6A4, HTR2A, and GRIK4) and outcomes,17 including some large clinical trials that conducted genome-wide association studies18-20 and meta-analyses across multiple studies.21,22 Most pharmacogenetic studies in psychiatric patients are small, and very few have included pediatric patients. However, with more interest in neuropsychiatric pharmacogenetics, these studies are becoming more common.23-26
Continue to: Limited evidence from studies of commercially available tests
Limited evidence from studies of commercially available tests
Several pharmacogenetic tests are commercially available, including some that focus on providing information that can be used specifically when prescribing psychiatric medications, such as the GeneSight Psychotropic test, CNSdose, Genomind, and Neuropharmagen.
In an industry-sponsored, nonrandomized clinical trial that included patients for whom prescribing decisions were made based on the GeneSight test, outcomes in adults were improved compared with treatment as usual,27 inpatient stays were shorter,28 and pharmacy costs were reduced.29 In one of these studies, the authors noted that the traditional, single-gene analysis was not associated with improved outcomes, whereas the multiple gene combination (pharmacokinetic and pharmacodynamic genes) was associated with improved outcomes among patients with depression.27 However, when GeneSightwas compared with treatment as usual in a small randomized trial, there was not a significant association between use of the test and improved outcomes among patients with treatment-resistant depression.30 The results of a much larger randomized trial (N = 1,167) are available31 and expected to be published, but patients younger than age 18 were excluded from this study.32 A retrospective study conducted in adult psychiatric patients found that patients whose treatment followed recommendations of a pharmacogenetic test including 20 genes were almost 4 times more likely to improve than patients whose treatment did not follow the recommendations.33
Pharmacogenetic testing at our pediatric inpatient unit
The Cincinnati Children’s Division of Child and Adolescent Psychiatry is the largest psychiatric inpatient service in a U.S. pediatric hospital. Starting in 2004, we adopted pharmacogenetically-guided dosing of psychiatric medications.34 CYP2D6 and CYP2C19 were chosen for testing because the enzymes encoded by these genes metabolize many of the antidepressants and antipsychotics that patients admitted to our unit will receive, and the clinicians wanted all available tools to help improve the care of these patients. To date, the Genetic Pharmacology Service (GPS) has performed >25,000 tests for variants in CYP2D6 and CYP2C19 as part of inpatient care. Patients provide a specimen (blood or buccal swab) at the time of admission to inpatient psychiatry, genotyping is performed onsite by the Molecular Genetics Laboratory (certified by the College of American Pathologists [CAP]/Clinical Laboratory Improvement Amendments [CLIA]) and the results are posted to the medical record within 2 business days. The report contains the patient’s alleles for CYP2D6 and CYP2C19, the genotype-predicted metabolizer phenotype, and dosing recommendations for 19 drugs (provided as a percentage of the standard dose). Insurance is billed for the test, and reimbursement is usually received when the test is performed as part of an inpatient stay.
The GPS team performed a retrospective chart review after the first panel was implemented in 2005.23 The study included 279 patients who were receiving a medication metabolized by one of the 2 genes tested. The poor metabolizers had the highest efficacy and highest number of adverse drug reactions, while ultra-rapid metabolizers had the lowest efficacy and lowest number of adverse reactions during their initial inpatient stay. In patients not treated with medications metabolized by CYP2D6 or CYP2C19, there was no association between metabolizer status and efficacy or adverse drug reactions. In this retrospective study, there was no association between metabolizer status and length of stay.
Overcoming the challenges
One challenge with many of the pharmacogenetic tests is interpretation of the results. The reports can span more than 20 pages, and clinicians may not have time to thoroughly read and understand how best to use all of this information. Sometimes the reports can make it seem like the first-line medication for the patient’s condition is not the best choice, but it could work well when dosed appropriately based on the patient’s genotype. Each commercially available test has a different way of presenting results,13 so when choosing a pharmacogenetic test, one should be sure to see a sample report. Vo et al35 recently reviewed factors to consider when choosing a pharmacogenetic test.
Continue to: Because patients and families also have difficulty understanding the reports...
Because patients and families also have difficulty understanding the reports, we created patient education sheets,36 written at an eighth grade level with feedback from parents and modeled on those provided by St. Jude Children’s Research Hospital.37 St. Jude Children’s Research Hospital also has pharmacogenetic competencies that pharmacists and nurses must pass.38,39 The following is a sample explanation that one of our nurses uses to educate parents on what is being tested and what effect the results will have on the treatment plan.
“During your child’s stay we will be completing a genetic test to help us understand how he/she processes the types of medications that we may be likely to start during their hospitalization. This does not tell us which medication will be best—unfortunately within the field of psychiatry there is still some unavoidable trial and error; rather, what it will do is tell us how to make sure that the dosing is at a level that would be safe for the way your child’s body breaks down the medicine, so that he/she can get the intended benefit of the medicine’s effects, while decreasing the risk of uncomfortable side effects, where possible.”
Other challenges in pharmacogenetic testing are the cost, disease risk, and concern about how genetic information will be used. Because these tests are often not covered by health insurance, some commercial pharmacogenetic testing companies offer an out-of-pocket maximum in the $250 to $350 range to reduce the cost to the patient. Some pharmacogenetic testing companies also test for genes associated with disease, so if a clinician orders the test, he or she may be responsible for sharing that information with the patient. For most pharmacogenetic testing companies, the turn-around time is 2 to 10 days. Genetic information is protected by federal laws, including Genetic Information Nondiscrimination Act (GINA) and Health Insurance Portability and Accountability Act (HIPAA).
The choice of psychotropic medication is complex, and although we would like pharmacogenetics to be the only answer to why every patient does or does not respond to a medication, it is not. Response to medication is influenced by age, comorbidities, illness severity, illness duration, compliance, gender, concomitant medications, and potentially more.40 Pharmacogenetics is another tool at the clinician’s disposal to help in choosing a medication and dose. There is a clear association between CYP2D6 and CYP2C19 and exposure to many antidepressants and antipsychotics (reviewed by Stingl et al3); however, the link between exposure and response is much weaker. It may be strengthened by the inclusion of pharmacodynamic information (the level of expression of the drug target), which can be influenced by genetic variants.41 At the present time, the most evidence exists for testing CYP2D6 and CYP2C19, and the CPIC4,5,15 and DWPG6 guidelines provide evidence-based recommendations for how to adjust medication dosages based on the results.
There is clearly much more research that needs to be done in the field of neuropsychiatric pharmacogenetics, especially in pediatric populations. As we see increased utilization of pharmacogenetic tests in psychiatry, there is also a need for pharmacogenetic education of patients, families, nurses, pharmacists, and psychiatrists. Several good pharmacogenetic resources that contain up-to-date summaries of the available evidence linking pharmacogenetic variants to medication response, implementation resources, and educational resources are available. These include CPIC (www.cpicpgx.org), PharmGKB (www.pharmgkb.org), and the IGNITE Spark Toolbox (https://ignite-genomics.org/spark-toolbox/clinicians/).
Acknowledgements
The author thanks Jen Milau, APRN, for the case study and sample explanation, and Jeffrey Strawn, MD, FAACP, Ethan Poweleit, and Stacey Aldrich, MS, for help with preparing this manuscript.
1. Cipriani A, Zhou X, Del Giovane C, et al. Comparative efficacy and tolerability of antidepressants for major depressive disorder in children and adolescents: a network meta-analysis. Lancet. 2016; 388(10047):881-890.
2. Correll CU, Sheridan EM, DelBello MP. Antipsychotic and mood stabilizer efficacy and tolerability in pediatric and adult patients with bipolar I mania: a comparative analysis of acute, randomized, placebo-controlled trials. Bipolar Disord. 2010;12(2):116-141.
3. Stingl JC, Brockmoller J, Viviani R. Genetic variability of drug-metabolizing enzymes: the dual impact on psychiatric therapy and regulation of brain function. Mol Psychiatry. 2013;18(3):273-287.
4. Hicks JK, Bishop JR, Sangkuhl K, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin Pharmacol Ther. 2015;98(2):127-134.
5. Hicks JK, Sangkuhl K, Swen JJ, et al. Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin Pharmacol Ther. 2017;102(1):37-44.
6. Swen JJ, Nijenhuis M, de Boer A, et al. Pharmacogenetics: from bench to byte--an update of guidelines. Clin Pharmacol Ther. 2011;89(5):662-673.
7. Swen JJ, Wilting I, de Goede AL, et al. Pharmacogenetics: from bench to byte. Clin Pharmacol Ther. 2008;83(5):781-787.
8. GENDEP Investigators, MARS Investigators, and STAR*D Investigators. Common genetic variation and antidepressant efficacy in major depressive disorder: a meta-analysis of three genome-wide pharmacogenetic studies. Am J Psychiatry. 2013;170(2):207-217.
9. Ji Y, Schaid DJ, Desta Z, et al. Citalopram and escitalopram plasma drug and metabolite concentrations: genome-wide associations. Br J Clin Pharmacol. 2014;78(2):373-383.
10. Werk AN, Cascorbi I. Functionalgene variants of CYP3A4. Clin Pharmacol Ther. 2014:96(3):340-348.
11. Pratt VM, Del Tredici AL, Hachad H, et al. Recommendations for clinical CYP2C19 genotyping allele selection: a report of the Association for Molecular Pathology. J Mol Diagn. 2018;20(3):269-276.
12. Bousman CA, Jaksa P, Pantelis C. Systematic evaluation of commercial pharmacogenetic testing in psychiatry: a focus on CYP2D6 and CYP2C19 allele coverage and results reporting. Pharmacogenet Genomics. 2017;27(11):387-393.
13. Hicks JK, Swen JJ, Gaedigk A. Challenges in CYP2D6 phenotype assignment from genotype data: a critical assessment and call for standardization. Curr Drug Metab. 2014;15(2):218-232.
14. Caudle KE, Klein TE, Hoffman JM, et al. Incorporation of pharmacogenomics into routine clinical practice: the Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline development process. Curr Drug Metab. 2014;15(2):209-217.
15. Hicks JK, Swen JJ, Thorn CF, et al. Clinical Pharmacogenetics Implementation Consortium guideline for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants. Clin Pharmacol Ther. 2013;93(5):402-408.
16. Quaranta S, Dupouey J, Colle R, et al. Pharmacogenetics of antidepressant drugs: State of the art and clinical implementation - recommendations from the French National Network of Pharmacogenetics. Therapie. 2017;72(2):311-318.
17. Fabbri C, Minarini A, Nitsu T, et al. Understanding the pharmacogenetics of selective serotonin reuptake inhibitors. Expert Opin Drug Metab Toxicol. 2014;10(8):1093-1118.
18. Mrazek DA, Rush AJ, Biernacka JM, et al. SLC6A4 variation and citalopram response. Am J Med Genet B Neuropsychiatr Genet. 2009;150B(3):341-351.
19. Biernacka JM, Sangkuhl K, Jenkins G, et al. The International SSRI Pharmacogenomics Consortium (ISPC): a genome-wide association study of antidepressant treatment response. Transl Psychiatry. 2015;5:e553. doi: 10.1038/tp.2015.47.
20. Horstmann S, Lucae S, Menke A, et al. Polymorphisms in GRIK4, HTR2A, and FKBP5 show interactive effects in predicting remission to antidepressant treatment. Neuropsychopharmacology. 2010;35(3):727-740.
21. Porcelli S, Fabbri C, Serretti A. Meta-analysis of serotonin transporter gene promoter polymorphism (5-HTTLPR) association with antidepressant efficacy. Eur Neuropsychopharmacol. 2012;22(4):239-258.
22. Niitsu T, Fabbri C, Bentini F, et al. Pharmacogenetics in major depression: a comprehensive meta-analysis. Prog Neuropsychopharmacol Biol Psychiatry. 2013;45:183-194.
23. Prows CA, Nick TG, Saldaña SN, et al. Drug-metabolizing enzyme genotypes and aggressive behavior treatment response in hospitalized pediatric psychiatric patients. J Child Adolesc Psychopharmacol. 2009;19(4):385-394.
24. Rotberg B, Kronenberg S, Carmel M, et al. Additive effects of 5-HTTLPR (serotonin transporter) and tryptophan hydroxylase 2 G-703T gene polymorphisms on the clinical response to citalopram among children and adolescents with depression and anxiety disorders. J Child Adolesc Psychopharmacol. 2013;23(2):117-122.
25. Kronenberg S, Apter A, Brent D, et al. Serotonin transporter polymorphism (5-HTTLPR) and citalopram effectiveness and side effects in children with depression and/or anxiety disorders. J Child Adolesc Psychopharmacol. 2007;17(6):741-750.
26. AlOlaby RR, Sweha SR, Silva M, et al. Molecular biomarkers predictive of sertraline treatment response in young children with fragile X syndrome. Brain Dev. 2017;39(6):483-492.
27. Altar CA, Carhart JM, Allen JD, et al. Clinical validity: Combinatorial pharmacogenomics predicts antidepressant responses and healthcare utilizations better than single gene phenotypes. Pharmacogenomics J. 2015;15(5):443-451.
28. Winner J, Allen JD, Altar CA, et al. Psychiatric pharmacogenomics predicts health resource utilization of outpatients with anxiety and depression. Transl Psychiatry. 2013;3:e242. doi:10.1038/tp.2013.2.
29. Winner JG, Carhart JM, Altar CA, et al. Combinatorial pharmacogenomic guidance for psychiatric medications reduces overall pharmacy costs in a 1 year prospective evaluation. Curr Med Res Opin. 2015;31(9):1633-1643.
30. Winner JG, Carhart JM, Altar CA, et al. A prospective, randomized, double-blind study assessing the clinical impact of integrated pharmacogenomic testing for major depressive disorder. Discov Med. 2013;16(89):219-227.
31. Genesight. GUIDED clinical study. https://genesight.com/greden-study/. Updated May 31, 2018. Accessed August 1, 2018.
32. U.S. National Library of Medicine ClinicalTrials.gov. Genomics used to improve DEpression decisions (GUIDED). https://clinicaltrials.gov/ct2/show/NCT02109939. Accessed July 24, 2018.
33. Espadaler J, Tuson M, Lopez-Ibor JM, et al. Pharmacogenetic testing for the guidance of psychiatric treatment: a multicenter retrospective analysis. CNS Spectrums. 2017;22(4):315-324.
34. Ramsey LB, Prows CA, Zhang K, et al. Implementation of pharmacogenetics at Cincinnati Children’s Hospital Medical Center: lessons learned over 14 years of personalizing medicine. Clin Pharmacol Ther. 2018. doi: 10.1002/cpt.1165. [Epub ahead of print].
35. Vo TT, Bell GC, Owusu Obeng A, et al. Pharmacogenomics implementation: considerations for selecting a reference laboratory. Pharmacotherapy. 2017;37(9):1014-1022.
36. Cincinnati Children’s Hospital. Genetic Pharmacology Service: Education. www.cincinnatichildrens.org/gpsinfo. Accessed August 1, 2018.
37. St. Jude Children’s Research Hospital. Do You Know...Cytochrome P450 2D6 (CYP2D6) and medicines. https://www.stjude.org/treatment/patient-resources/caregiver-resources/patient-family-education-sheets/pharmacy-and-medicines/cytochrome-p450-2d6-cyp2d6-and-medicines.html. Accessed August 1, 2018.
38. St. Jude Children’s Research Hospital. Implementation Resources for Professionals: Clinical Pharmacogenetics at St. Jude. https://www.stjude.org/research/clinical-trials/pg4kds-pharmaceutical-science/implementation-resources-for-professionals.html. Accessed August 1, 2018.
39. Hoffman JM, Haider CE, Wilkinson MR, et al. PG4KDS: a model for the clinical implementation of pre-emptive pharmacogenetics. Am J Med Genet C Semin Med Genet. 2014;166C(1):45-55.
40. Wehry AM, Ramsey LB, Dulemba SE, et al. Pharmacogenomic testing in child and adolescent psychiatry: an evidence-based review. Curr Probl Pediatr Adolesc Health Care. 2018;48(2):40-49.
41. Tomita T, Yasui-Furukori N, Nakagami T, et al. The influence of 5-HTTLPR genotype on the association between the plasma concentration and therapeutic effect of paroxetine in patients with major depressive disorder. PLoS One. 2014;9(5):e98099. doi: 10.1371/journal.pone.0098099.
1. Cipriani A, Zhou X, Del Giovane C, et al. Comparative efficacy and tolerability of antidepressants for major depressive disorder in children and adolescents: a network meta-analysis. Lancet. 2016; 388(10047):881-890.
2. Correll CU, Sheridan EM, DelBello MP. Antipsychotic and mood stabilizer efficacy and tolerability in pediatric and adult patients with bipolar I mania: a comparative analysis of acute, randomized, placebo-controlled trials. Bipolar Disord. 2010;12(2):116-141.
3. Stingl JC, Brockmoller J, Viviani R. Genetic variability of drug-metabolizing enzymes: the dual impact on psychiatric therapy and regulation of brain function. Mol Psychiatry. 2013;18(3):273-287.
4. Hicks JK, Bishop JR, Sangkuhl K, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin Pharmacol Ther. 2015;98(2):127-134.
5. Hicks JK, Sangkuhl K, Swen JJ, et al. Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin Pharmacol Ther. 2017;102(1):37-44.
6. Swen JJ, Nijenhuis M, de Boer A, et al. Pharmacogenetics: from bench to byte--an update of guidelines. Clin Pharmacol Ther. 2011;89(5):662-673.
7. Swen JJ, Wilting I, de Goede AL, et al. Pharmacogenetics: from bench to byte. Clin Pharmacol Ther. 2008;83(5):781-787.
8. GENDEP Investigators, MARS Investigators, and STAR*D Investigators. Common genetic variation and antidepressant efficacy in major depressive disorder: a meta-analysis of three genome-wide pharmacogenetic studies. Am J Psychiatry. 2013;170(2):207-217.
9. Ji Y, Schaid DJ, Desta Z, et al. Citalopram and escitalopram plasma drug and metabolite concentrations: genome-wide associations. Br J Clin Pharmacol. 2014;78(2):373-383.
10. Werk AN, Cascorbi I. Functionalgene variants of CYP3A4. Clin Pharmacol Ther. 2014:96(3):340-348.
11. Pratt VM, Del Tredici AL, Hachad H, et al. Recommendations for clinical CYP2C19 genotyping allele selection: a report of the Association for Molecular Pathology. J Mol Diagn. 2018;20(3):269-276.
12. Bousman CA, Jaksa P, Pantelis C. Systematic evaluation of commercial pharmacogenetic testing in psychiatry: a focus on CYP2D6 and CYP2C19 allele coverage and results reporting. Pharmacogenet Genomics. 2017;27(11):387-393.
13. Hicks JK, Swen JJ, Gaedigk A. Challenges in CYP2D6 phenotype assignment from genotype data: a critical assessment and call for standardization. Curr Drug Metab. 2014;15(2):218-232.
14. Caudle KE, Klein TE, Hoffman JM, et al. Incorporation of pharmacogenomics into routine clinical practice: the Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline development process. Curr Drug Metab. 2014;15(2):209-217.
15. Hicks JK, Swen JJ, Thorn CF, et al. Clinical Pharmacogenetics Implementation Consortium guideline for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants. Clin Pharmacol Ther. 2013;93(5):402-408.
16. Quaranta S, Dupouey J, Colle R, et al. Pharmacogenetics of antidepressant drugs: State of the art and clinical implementation - recommendations from the French National Network of Pharmacogenetics. Therapie. 2017;72(2):311-318.
17. Fabbri C, Minarini A, Nitsu T, et al. Understanding the pharmacogenetics of selective serotonin reuptake inhibitors. Expert Opin Drug Metab Toxicol. 2014;10(8):1093-1118.
18. Mrazek DA, Rush AJ, Biernacka JM, et al. SLC6A4 variation and citalopram response. Am J Med Genet B Neuropsychiatr Genet. 2009;150B(3):341-351.
19. Biernacka JM, Sangkuhl K, Jenkins G, et al. The International SSRI Pharmacogenomics Consortium (ISPC): a genome-wide association study of antidepressant treatment response. Transl Psychiatry. 2015;5:e553. doi: 10.1038/tp.2015.47.
20. Horstmann S, Lucae S, Menke A, et al. Polymorphisms in GRIK4, HTR2A, and FKBP5 show interactive effects in predicting remission to antidepressant treatment. Neuropsychopharmacology. 2010;35(3):727-740.
21. Porcelli S, Fabbri C, Serretti A. Meta-analysis of serotonin transporter gene promoter polymorphism (5-HTTLPR) association with antidepressant efficacy. Eur Neuropsychopharmacol. 2012;22(4):239-258.
22. Niitsu T, Fabbri C, Bentini F, et al. Pharmacogenetics in major depression: a comprehensive meta-analysis. Prog Neuropsychopharmacol Biol Psychiatry. 2013;45:183-194.
23. Prows CA, Nick TG, Saldaña SN, et al. Drug-metabolizing enzyme genotypes and aggressive behavior treatment response in hospitalized pediatric psychiatric patients. J Child Adolesc Psychopharmacol. 2009;19(4):385-394.
24. Rotberg B, Kronenberg S, Carmel M, et al. Additive effects of 5-HTTLPR (serotonin transporter) and tryptophan hydroxylase 2 G-703T gene polymorphisms on the clinical response to citalopram among children and adolescents with depression and anxiety disorders. J Child Adolesc Psychopharmacol. 2013;23(2):117-122.
25. Kronenberg S, Apter A, Brent D, et al. Serotonin transporter polymorphism (5-HTTLPR) and citalopram effectiveness and side effects in children with depression and/or anxiety disorders. J Child Adolesc Psychopharmacol. 2007;17(6):741-750.
26. AlOlaby RR, Sweha SR, Silva M, et al. Molecular biomarkers predictive of sertraline treatment response in young children with fragile X syndrome. Brain Dev. 2017;39(6):483-492.
27. Altar CA, Carhart JM, Allen JD, et al. Clinical validity: Combinatorial pharmacogenomics predicts antidepressant responses and healthcare utilizations better than single gene phenotypes. Pharmacogenomics J. 2015;15(5):443-451.
28. Winner J, Allen JD, Altar CA, et al. Psychiatric pharmacogenomics predicts health resource utilization of outpatients with anxiety and depression. Transl Psychiatry. 2013;3:e242. doi:10.1038/tp.2013.2.
29. Winner JG, Carhart JM, Altar CA, et al. Combinatorial pharmacogenomic guidance for psychiatric medications reduces overall pharmacy costs in a 1 year prospective evaluation. Curr Med Res Opin. 2015;31(9):1633-1643.
30. Winner JG, Carhart JM, Altar CA, et al. A prospective, randomized, double-blind study assessing the clinical impact of integrated pharmacogenomic testing for major depressive disorder. Discov Med. 2013;16(89):219-227.
31. Genesight. GUIDED clinical study. https://genesight.com/greden-study/. Updated May 31, 2018. Accessed August 1, 2018.
32. U.S. National Library of Medicine ClinicalTrials.gov. Genomics used to improve DEpression decisions (GUIDED). https://clinicaltrials.gov/ct2/show/NCT02109939. Accessed July 24, 2018.
33. Espadaler J, Tuson M, Lopez-Ibor JM, et al. Pharmacogenetic testing for the guidance of psychiatric treatment: a multicenter retrospective analysis. CNS Spectrums. 2017;22(4):315-324.
34. Ramsey LB, Prows CA, Zhang K, et al. Implementation of pharmacogenetics at Cincinnati Children’s Hospital Medical Center: lessons learned over 14 years of personalizing medicine. Clin Pharmacol Ther. 2018. doi: 10.1002/cpt.1165. [Epub ahead of print].
35. Vo TT, Bell GC, Owusu Obeng A, et al. Pharmacogenomics implementation: considerations for selecting a reference laboratory. Pharmacotherapy. 2017;37(9):1014-1022.
36. Cincinnati Children’s Hospital. Genetic Pharmacology Service: Education. www.cincinnatichildrens.org/gpsinfo. Accessed August 1, 2018.
37. St. Jude Children’s Research Hospital. Do You Know...Cytochrome P450 2D6 (CYP2D6) and medicines. https://www.stjude.org/treatment/patient-resources/caregiver-resources/patient-family-education-sheets/pharmacy-and-medicines/cytochrome-p450-2d6-cyp2d6-and-medicines.html. Accessed August 1, 2018.
38. St. Jude Children’s Research Hospital. Implementation Resources for Professionals: Clinical Pharmacogenetics at St. Jude. https://www.stjude.org/research/clinical-trials/pg4kds-pharmaceutical-science/implementation-resources-for-professionals.html. Accessed August 1, 2018.
39. Hoffman JM, Haider CE, Wilkinson MR, et al. PG4KDS: a model for the clinical implementation of pre-emptive pharmacogenetics. Am J Med Genet C Semin Med Genet. 2014;166C(1):45-55.
40. Wehry AM, Ramsey LB, Dulemba SE, et al. Pharmacogenomic testing in child and adolescent psychiatry: an evidence-based review. Curr Probl Pediatr Adolesc Health Care. 2018;48(2):40-49.
41. Tomita T, Yasui-Furukori N, Nakagami T, et al. The influence of 5-HTTLPR genotype on the association between the plasma concentration and therapeutic effect of paroxetine in patients with major depressive disorder. PLoS One. 2014;9(5):e98099. doi: 10.1371/journal.pone.0098099.
