User login
Impact of Rapid Blood Culture Identification on Antibiotic De-escalation at a Veterans Affairs Medical Center
About 530,000 to 628,000 episodes of bloodstream infections (BSI) occur annually in the US.1 Early identification and treatment of bacteremia are essential to improve patient outcomes because it allows for more timely targeted antibiotic therapy.2 Organism identification and susceptibility testing can take 2 to 5 days, prolonging the use of broad-spectrum empiric antibiotics and increasing the risk of adverse events.3,4 The Infectious Disease Society of America recommends the use of rapid diagnostic testing and antimicrobial stewardship programs (ASPs) to improve rates of antibiotic susceptibilities to targeted antibiotics and optimize resource utilization.3 Rapid blood culture identification (BCID) technologies reduce the duration of empiric antibiotics in patients with contaminated blood cultures, resulting in shorter hospital stays and saving money per each patient tested.4
In March 2023, Veteran Health Indiana (VHI) implemented the BioFire FilmArray Blood Culture Identification (BCID2), a BSI panel test that identifies select gram-negative bacteria, gram-positive bacteria, yeast, and antimicrobial resistance genes with an aggregate sensitivity of 99% and a specificity of 99.8%. The BCID2 presents clinically relevant information faster than traditional culture methods, allowing clinicians to make more efficient and educated antibiotic regimen decisions than with previous methods.5
It takes 24 to 48 hours from blood collection for culture incubation, positivity, and gram staining to occur at VHI. If the gram stain is positive, the blood culture is placed on the BioFire BCID2 in addition to traditional culture medium. BioFire BCID2 results are ready in 45 to 60 minutes. Results are uploaded into the electronic health record (EHR) ≤ 2 hours after they are obtained and the primary team is notified if the test is positive for certain critical results. Susceptibility testing of an identified organism typically requires an additional 24 to 48 hours for finalization. VHI Infectious Disease created an evidence-based antibiotic recommendation chart for certain medication(s) and alternate therapies based on the reported organism and its interpreted presence of resistance markers (eg, ceftriaxone for Escherichia coli when extended-spectrum beta lactamases are not detected vs meropenem if extended-spectrum beta lactamases marker are present). These charts optimize the antibiotic regimen while awaiting susceptibility finalizations.
Two previous studies describe the impact of rapid diagnostic testing technology at US Department of Veterans Affairs (VA) medical centers.6,7 In Texas, the ASP reviewed BCID panel results via clinical decision support software for about 1 hour per day.6 A Los Angeles study analyzed the impact of Biofire BCID with an interpretation guide centered on unnecessary vancomycin use and determined that shorter duration of the medication may have been the result of more frequent infectious disease consultation.7
This study assessed the time to optimal antibiotic de-escalation before and after the implementation of BioFire BCID2 with results reviewed by the ASP without active notification or assistance of any clinical decision support technology. The primary objective was to evaluate difference in time to optimal antibiotics from blood culture draw pre- vs postintervention. Secondary objectives included differences in time to organism identification, difference in time on broad-spectrum antibiotics, and difference in time to appropriate antibiotics.
Methods
This quasi-experimental retrospective chart review assessed the impact of BioFire BCID2 use on timely antibiotic de-escalation for patients who experienced a BSI at VHI between March 1, 2022, and October 1, 2023. Microbiology laboratory records identified eligible patients with positive blood cultures within the study time frame. Data were collected from the VHI EHR.
Patients were included if they had a positive bacterial blood culture and received ≥ 1 antibiotic indicated for bacteremia while receiving inpatient care. Patients were excluded if they died prior to blood culture results, transferred out of VHI, left against medical advice, or had untreated contaminants in blood culture results (ie, never received antibiotics aimed at the contaminated culture).
Patient lists were generated for before and after implementation of BioFire BCID2 (pre- and postintervention) using the VHI EHR and microbiology laboratory record system. The pre- and postinterventions groups were different sizes. As a result, a random sampling of the preintervention group was selected and included patients from March 1, 2022, through March 26, 2023. The postintervention group was smaller due to time constraints between initiation of BioFire BCID2 for data collection and included all patients from March 27, 2023, through October 1, 2023.
Optimal antibiotics were defined as escalation from inappropriate therapy to broader agent(s), de-escalation from broad-spectrum therapy to targeted agent(s), discontinuation of therapy due to an organism being identified as a contaminant, or optimization of a regimen to the preferred antimicrobial agent based on evidence-based consensus guidelines. Broad-spectrum antibiotics included: piperacillin/tazobactam, cefepime, ceftazidime, ceftazidime-avibactam, cefiderocol, carbapenems, fluroquinolones, vancomycin, daptomycin, ceftaroline, linezolid, or aztreonam. Appropriate antibiotics were defined as those with activity toward the final identified organism(s).
Deidentified participant data were entered into Microsoft Excel and kept on a secure VA server to complete statistical analyses. Parametric continuous data, such as age, were analyzed using the t-test, while nonparametric continuous data, such as time to optimal antibiotics, were analyzed using the Mann-Whitney U test. Categorical data, like sex and race, were analyzed using either Fisher exact test for small sample sizes or X2 test for a larger sample size. Statistical significance levels was defined as P < .05.
Results
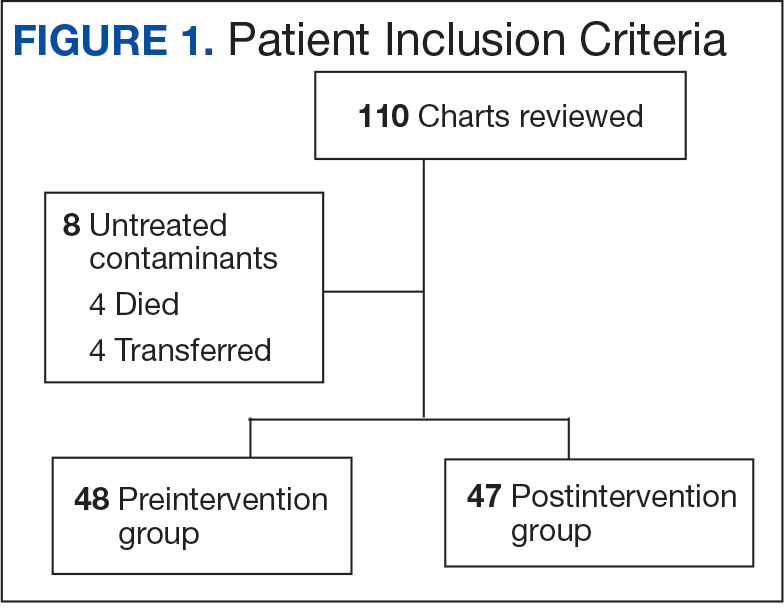
Using patient lists drawn from the EHR and the microbiology laboratory records, 110 electronic charts were randomly selected for review. Fifteen patients were excluded: 8 had untreated contaminants, 4 died, and 3 were transferred out of VHI. Of the 95 patients included, 48 were in the preintervention group and 47 were in the postintervention group (Figure 1).
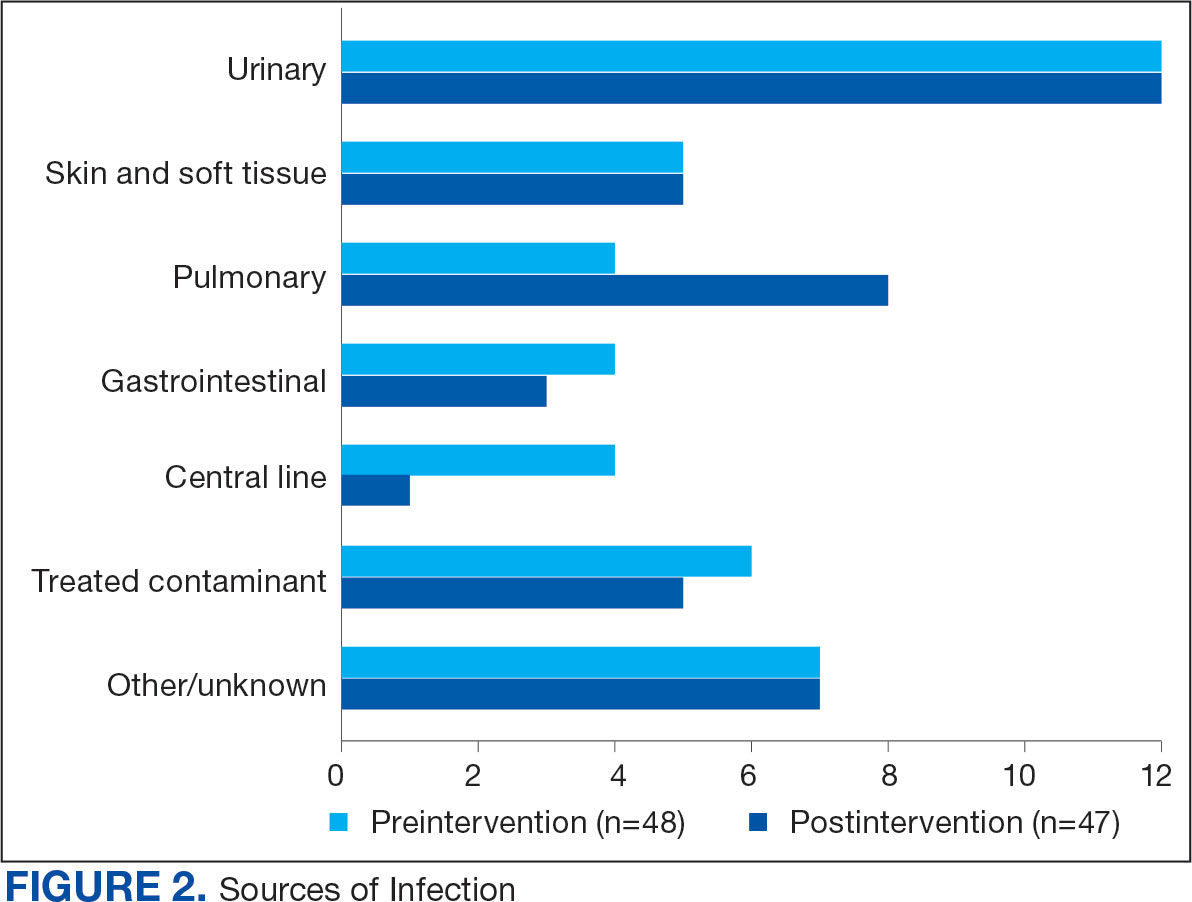
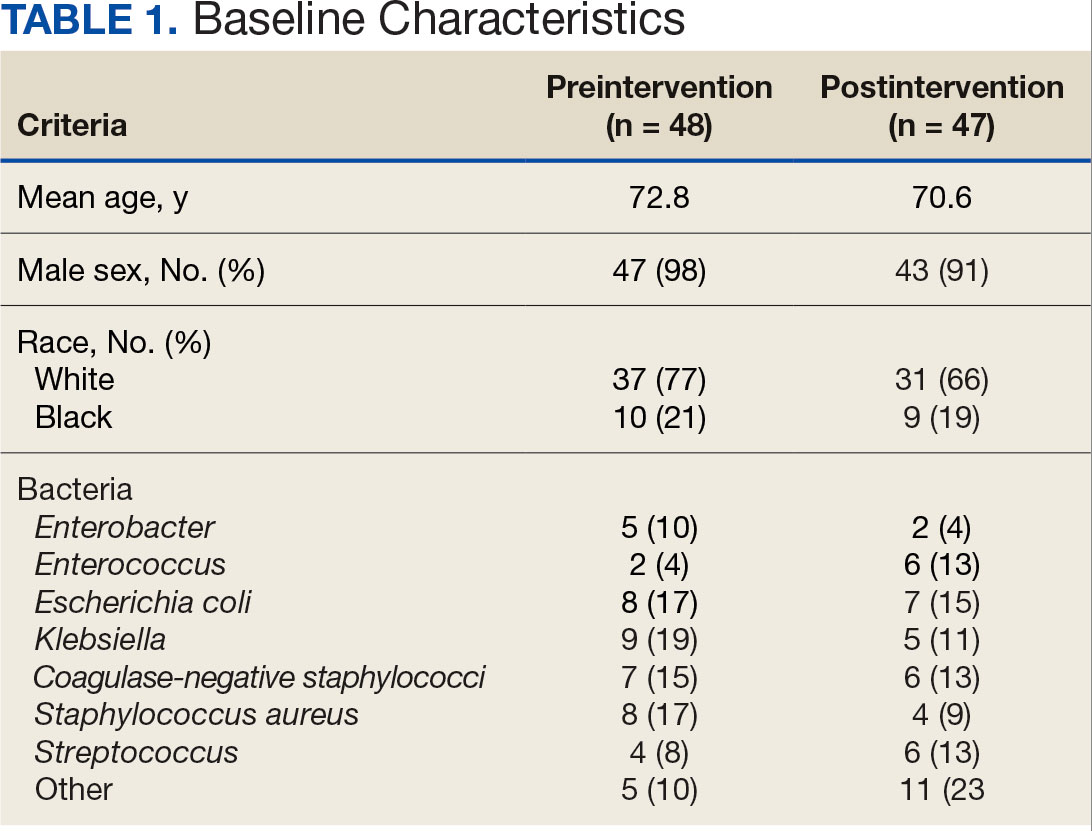
Baseline characteristics were similar between the 2 groups (Table 1). Most patients were White males aged > 70 years in the EHR. The urinary tract was the most common source of infection, impacting 12 patients in each group (Figure 2). Escherichia coli, Klebsiella, Staphylococcus, and Streptococcus were the most common bloodstream isolates identified.
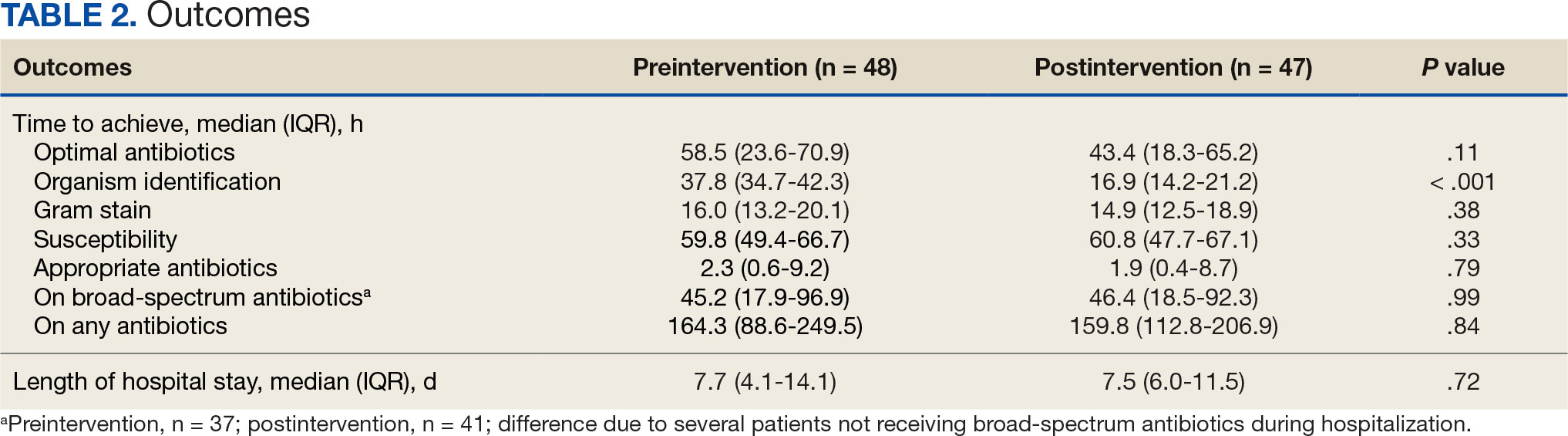
The median time to optimal antibiotics in the preintervention group was 58.5 hours vs 43.4 hours in the postintervention group (P = .11). The median time to organism identification was 37.8 hours in the preintervention group vs 16.9 hours in the postintervention group (P < .001). The median time on broad-spectrum antibiotics was 45.2 hours in the preintervention group vs 46.6 hours in the postintervention group (P = .99). The median time on appropriate antibiotics in the preintervention group was 2.3 hours vs 1.9 hours in the postintervention group (P = .79). Differences in other measured outcomes between the groups were not statistically significant (Table 2).
Although implementation of rapid diagnostic technology reduced the median time to optimal antibiotics, the results were not statistically significant. Shorter time to organism identification in the postintervention group compared to the preintervention group was the lone statistically significant metric (P < .001).
Discussion
A lack of statistical significance in the primary outcome may have been due to nonadherence to facility de-escalation protocols or a suboptimal BioFire BCID2 result notification system. Additionally, use of rapid BCID at VHI may improve over time as clinicians become more familiar with the technology. Gaps in clinical pharmacy coverage during the night shift may have also contributed to delays in antibiotic optimization, particularly if other clinicians are not equipped with the knowledge or training to appropriately deescalate antibiotics based on microorganisms identified. A 2017 study by Donner et al concluded that physician interpretation of BCID results is suboptimal and should be augmented with clinical decision support tools as new technology becomes available.8 Despite the statistically insignificant results of this study, it did highlight potential areas of improvement which can lead to improved patient care.
Previous research has evaluated the impact of rapid BCID technology on antibiotic treatment and clinical outcomes. Chiasson et al found that median time to optimal therapy was 73.8 hours in the pre-BCID arm compared to 34.7 hours in the post- BCID arm (P ≤ .001), emphasizing the importance of combining rapid BCID with clinical decision support tools and pharmacy input.6 Senok et al found that BCID2 implementation led to a significant decrease in median time to culture result, which informed optimal antibiotic therapy and decreased 30-day mortality in the intensive care setting.9 In contrast, the current study did not stratify patients according to medical ward or illness severity even though clinicians may be less likely to de-escalate antibiotic therapy in critically ill patients.
Bae et al reported findings consistent with the current study and concluded that BCID did not affect the clinical outcomes of overall BSIs; however, it contributed to early administration of effective antibiotics in cases of BSIs caused by multidrug-resistant organisms.10 Results of this study were not stratified according to multidrug-resistant organisms because the sample size was too small. The current study also included patients with polymicrobial infections, which may have impacted the results due to a less streamlined approach to antibiotic optimization.
Limitations
This single-center, retrospective study had a small sample size, short time frame, and lacked patient diversity, and therefore may not be generalizable to other health care systems. The sample size was limited by shorter date range and smaller patient list between BioFire BCID2 implementation and data collection, which was used to determine the number of charts selected in each group. Some patients received antibiotics prior to blood cultures being drawn, which may falsely decrease time to optimal/ appropriate antibiotics and falsely increase time on broad spectrum/any antibiotics due to early antibiotic administration. The total number of patients on broad-spectrum antibiotics differed from the total number of patients for other outcomes because several patients never received the defined broad spectrum antibiotics.
Conclusions
When combined with a pre-existing ASP without active notification, the implementation of BioFire BCID2 did not return statistically significant data showing a decrease in time to optimal antibiotics, time to appropriate antibiotics, or time on broad-spectrum antibiotics at VHI. To make this program more successful, pharmacist intervention and clinical decision support tools may be needed.
Additional research is required to determine the optimal integration of antimicrobial stewardship, rapid diagnostic technology, and pharmacy services for maximum benefit. Even though the primary outcome was not statistically significant, the results may be clinically significant from a stewardship perspective. Realigning microbiology workflows to mimic other research, which emphasizes the importance of funneling rapid BCID results through the ASP, may improve outcomes. Future studies may be warranted following the implementation of clinical decision support tools to assess their impact on stewardship practices and patient outcomes.
- Goto M, Al-Hasan MN. Overall burden of bloodstream infection and nosocomial bloodstream infection in North America and Europe. Clin Microbiol Infect. 2013;19(6):501- 509. doi:10.1111/1469-0691.12195
- Pardo J, Klinker KP, Borgert SJ, Butler BM, Giglio PG, Rand KH. Clinical and economic impact of antimicrobial stewardship interventions with the FilmArray blood culture identification panel. Diagn Microbiol Infect Dis. 2016;84(2):159-164. doi:10.1016/j.diagmicrobio.2015.10.023.
- Barlam TF, Cosgrove SE, Abbo LM, et al. Implementing an antibiotic stewardship program: guidelines by the Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America. Clin Infect Dis. 2016;62(10):e51-e77. doi:10.1093/cid/ciw118
- BIOFIRE® Blood Culture Identification 2 (BCID2) Panel. Biomerierux. Updated 2025. Accessed May 10, 2025. https://www.biofiredx.com/products/the-filmarray-panels/filmarraybcid/
- Huang AM, Newton D, Kunapuli A, et al. Impact of rapid organism identification via matrix-assisted laser desorption/ionization time-of-flight combined with antimicrobial stewardship team intervention in adult patients with bacteremia and candidemia. Clin Infect Dis. 2013;57(9):1237-1245. doi:10.1093/cid/cit498
- Chiasson JM, Smith WJ, Jodlowski TZ, Kouma MA, Cutrell JB. Impact of a rapid blood culture diagnostic panel on time to optimal antimicrobial therapy at a veterans affairs medical center. J Pharm Pract. 2022;35(5):722-729. doi:10.1177/08971900211000686
- Wu S, Watson RL, Graber CJ. 2007. Impact of combining rapid diagnostics with an interpretation guide on vancomycin usage for contaminant blood cultures growing coagulase- negative staphylococci (CoNS). Open Forum Infect Dis. 2019;6(Suppl 2):S674. doi:10.1093/ofid/ofz360.1687
- Donner LM, Campbell WS, Lyden E, Van Schooneveld TC. Assessment of rapid-blood-culture-identification result interpretation and antibiotic prescribing practices. J Clin Microbiol. 2017;55(5):1496-1507. doi:10.1128/JCM.02395-16
- Senok A, Dabal LA, Alfaresi M, et al. Clinical impact of the BIOFIRE blood culture identification 2 panel in adult patients with bloodstream infection: a multicentre observational study in the United Arab Emirates. Diagnostics (Basel). 2023;13(14):2433. doi:10.3390/diagnostics13142433
- Bae JY, Bae J, So MK, Choi HJ, Lee M. The impact of the rapid blood culture identification panel on antibiotic treatment and clinical outcomes in bloodstream infections, particularly those associated with multidrug-resistant micro-organisms. Diagnostics (Basel). 2023;13(23):3504. doi:10.3390/diagnostics13233504
About 530,000 to 628,000 episodes of bloodstream infections (BSI) occur annually in the US.1 Early identification and treatment of bacteremia are essential to improve patient outcomes because it allows for more timely targeted antibiotic therapy.2 Organism identification and susceptibility testing can take 2 to 5 days, prolonging the use of broad-spectrum empiric antibiotics and increasing the risk of adverse events.3,4 The Infectious Disease Society of America recommends the use of rapid diagnostic testing and antimicrobial stewardship programs (ASPs) to improve rates of antibiotic susceptibilities to targeted antibiotics and optimize resource utilization.3 Rapid blood culture identification (BCID) technologies reduce the duration of empiric antibiotics in patients with contaminated blood cultures, resulting in shorter hospital stays and saving money per each patient tested.4
In March 2023, Veteran Health Indiana (VHI) implemented the BioFire FilmArray Blood Culture Identification (BCID2), a BSI panel test that identifies select gram-negative bacteria, gram-positive bacteria, yeast, and antimicrobial resistance genes with an aggregate sensitivity of 99% and a specificity of 99.8%. The BCID2 presents clinically relevant information faster than traditional culture methods, allowing clinicians to make more efficient and educated antibiotic regimen decisions than with previous methods.5
It takes 24 to 48 hours from blood collection for culture incubation, positivity, and gram staining to occur at VHI. If the gram stain is positive, the blood culture is placed on the BioFire BCID2 in addition to traditional culture medium. BioFire BCID2 results are ready in 45 to 60 minutes. Results are uploaded into the electronic health record (EHR) ≤ 2 hours after they are obtained and the primary team is notified if the test is positive for certain critical results. Susceptibility testing of an identified organism typically requires an additional 24 to 48 hours for finalization. VHI Infectious Disease created an evidence-based antibiotic recommendation chart for certain medication(s) and alternate therapies based on the reported organism and its interpreted presence of resistance markers (eg, ceftriaxone for Escherichia coli when extended-spectrum beta lactamases are not detected vs meropenem if extended-spectrum beta lactamases marker are present). These charts optimize the antibiotic regimen while awaiting susceptibility finalizations.
Two previous studies describe the impact of rapid diagnostic testing technology at US Department of Veterans Affairs (VA) medical centers.6,7 In Texas, the ASP reviewed BCID panel results via clinical decision support software for about 1 hour per day.6 A Los Angeles study analyzed the impact of Biofire BCID with an interpretation guide centered on unnecessary vancomycin use and determined that shorter duration of the medication may have been the result of more frequent infectious disease consultation.7
This study assessed the time to optimal antibiotic de-escalation before and after the implementation of BioFire BCID2 with results reviewed by the ASP without active notification or assistance of any clinical decision support technology. The primary objective was to evaluate difference in time to optimal antibiotics from blood culture draw pre- vs postintervention. Secondary objectives included differences in time to organism identification, difference in time on broad-spectrum antibiotics, and difference in time to appropriate antibiotics.
Methods
This quasi-experimental retrospective chart review assessed the impact of BioFire BCID2 use on timely antibiotic de-escalation for patients who experienced a BSI at VHI between March 1, 2022, and October 1, 2023. Microbiology laboratory records identified eligible patients with positive blood cultures within the study time frame. Data were collected from the VHI EHR.
Patients were included if they had a positive bacterial blood culture and received ≥ 1 antibiotic indicated for bacteremia while receiving inpatient care. Patients were excluded if they died prior to blood culture results, transferred out of VHI, left against medical advice, or had untreated contaminants in blood culture results (ie, never received antibiotics aimed at the contaminated culture).
Patient lists were generated for before and after implementation of BioFire BCID2 (pre- and postintervention) using the VHI EHR and microbiology laboratory record system. The pre- and postinterventions groups were different sizes. As a result, a random sampling of the preintervention group was selected and included patients from March 1, 2022, through March 26, 2023. The postintervention group was smaller due to time constraints between initiation of BioFire BCID2 for data collection and included all patients from March 27, 2023, through October 1, 2023.
Optimal antibiotics were defined as escalation from inappropriate therapy to broader agent(s), de-escalation from broad-spectrum therapy to targeted agent(s), discontinuation of therapy due to an organism being identified as a contaminant, or optimization of a regimen to the preferred antimicrobial agent based on evidence-based consensus guidelines. Broad-spectrum antibiotics included: piperacillin/tazobactam, cefepime, ceftazidime, ceftazidime-avibactam, cefiderocol, carbapenems, fluroquinolones, vancomycin, daptomycin, ceftaroline, linezolid, or aztreonam. Appropriate antibiotics were defined as those with activity toward the final identified organism(s).
Deidentified participant data were entered into Microsoft Excel and kept on a secure VA server to complete statistical analyses. Parametric continuous data, such as age, were analyzed using the t-test, while nonparametric continuous data, such as time to optimal antibiotics, were analyzed using the Mann-Whitney U test. Categorical data, like sex and race, were analyzed using either Fisher exact test for small sample sizes or X2 test for a larger sample size. Statistical significance levels was defined as P < .05.
Results
Using patient lists drawn from the EHR and the microbiology laboratory records, 110 electronic charts were randomly selected for review. Fifteen patients were excluded: 8 had untreated contaminants, 4 died, and 3 were transferred out of VHI. Of the 95 patients included, 48 were in the preintervention group and 47 were in the postintervention group (Figure 1).

Baseline characteristics were similar between the 2 groups (Table 1). Most patients were White males aged > 70 years in the EHR. The urinary tract was the most common source of infection, impacting 12 patients in each group (Figure 2). Escherichia coli, Klebsiella, Staphylococcus, and Streptococcus were the most common bloodstream isolates identified.


The median time to optimal antibiotics in the preintervention group was 58.5 hours vs 43.4 hours in the postintervention group (P = .11). The median time to organism identification was 37.8 hours in the preintervention group vs 16.9 hours in the postintervention group (P < .001). The median time on broad-spectrum antibiotics was 45.2 hours in the preintervention group vs 46.6 hours in the postintervention group (P = .99). The median time on appropriate antibiotics in the preintervention group was 2.3 hours vs 1.9 hours in the postintervention group (P = .79). Differences in other measured outcomes between the groups were not statistically significant (Table 2).

Although implementation of rapid diagnostic technology reduced the median time to optimal antibiotics, the results were not statistically significant. Shorter time to organism identification in the postintervention group compared to the preintervention group was the lone statistically significant metric (P < .001).
Discussion
A lack of statistical significance in the primary outcome may have been due to nonadherence to facility de-escalation protocols or a suboptimal BioFire BCID2 result notification system. Additionally, use of rapid BCID at VHI may improve over time as clinicians become more familiar with the technology. Gaps in clinical pharmacy coverage during the night shift may have also contributed to delays in antibiotic optimization, particularly if other clinicians are not equipped with the knowledge or training to appropriately deescalate antibiotics based on microorganisms identified. A 2017 study by Donner et al concluded that physician interpretation of BCID results is suboptimal and should be augmented with clinical decision support tools as new technology becomes available.8 Despite the statistically insignificant results of this study, it did highlight potential areas of improvement which can lead to improved patient care.
Previous research has evaluated the impact of rapid BCID technology on antibiotic treatment and clinical outcomes. Chiasson et al found that median time to optimal therapy was 73.8 hours in the pre-BCID arm compared to 34.7 hours in the post- BCID arm (P ≤ .001), emphasizing the importance of combining rapid BCID with clinical decision support tools and pharmacy input.6 Senok et al found that BCID2 implementation led to a significant decrease in median time to culture result, which informed optimal antibiotic therapy and decreased 30-day mortality in the intensive care setting.9 In contrast, the current study did not stratify patients according to medical ward or illness severity even though clinicians may be less likely to de-escalate antibiotic therapy in critically ill patients.
Bae et al reported findings consistent with the current study and concluded that BCID did not affect the clinical outcomes of overall BSIs; however, it contributed to early administration of effective antibiotics in cases of BSIs caused by multidrug-resistant organisms.10 Results of this study were not stratified according to multidrug-resistant organisms because the sample size was too small. The current study also included patients with polymicrobial infections, which may have impacted the results due to a less streamlined approach to antibiotic optimization.
Limitations
This single-center, retrospective study had a small sample size, short time frame, and lacked patient diversity, and therefore may not be generalizable to other health care systems. The sample size was limited by shorter date range and smaller patient list between BioFire BCID2 implementation and data collection, which was used to determine the number of charts selected in each group. Some patients received antibiotics prior to blood cultures being drawn, which may falsely decrease time to optimal/ appropriate antibiotics and falsely increase time on broad spectrum/any antibiotics due to early antibiotic administration. The total number of patients on broad-spectrum antibiotics differed from the total number of patients for other outcomes because several patients never received the defined broad spectrum antibiotics.
Conclusions
When combined with a pre-existing ASP without active notification, the implementation of BioFire BCID2 did not return statistically significant data showing a decrease in time to optimal antibiotics, time to appropriate antibiotics, or time on broad-spectrum antibiotics at VHI. To make this program more successful, pharmacist intervention and clinical decision support tools may be needed.
Additional research is required to determine the optimal integration of antimicrobial stewardship, rapid diagnostic technology, and pharmacy services for maximum benefit. Even though the primary outcome was not statistically significant, the results may be clinically significant from a stewardship perspective. Realigning microbiology workflows to mimic other research, which emphasizes the importance of funneling rapid BCID results through the ASP, may improve outcomes. Future studies may be warranted following the implementation of clinical decision support tools to assess their impact on stewardship practices and patient outcomes.
About 530,000 to 628,000 episodes of bloodstream infections (BSI) occur annually in the US.1 Early identification and treatment of bacteremia are essential to improve patient outcomes because it allows for more timely targeted antibiotic therapy.2 Organism identification and susceptibility testing can take 2 to 5 days, prolonging the use of broad-spectrum empiric antibiotics and increasing the risk of adverse events.3,4 The Infectious Disease Society of America recommends the use of rapid diagnostic testing and antimicrobial stewardship programs (ASPs) to improve rates of antibiotic susceptibilities to targeted antibiotics and optimize resource utilization.3 Rapid blood culture identification (BCID) technologies reduce the duration of empiric antibiotics in patients with contaminated blood cultures, resulting in shorter hospital stays and saving money per each patient tested.4
In March 2023, Veteran Health Indiana (VHI) implemented the BioFire FilmArray Blood Culture Identification (BCID2), a BSI panel test that identifies select gram-negative bacteria, gram-positive bacteria, yeast, and antimicrobial resistance genes with an aggregate sensitivity of 99% and a specificity of 99.8%. The BCID2 presents clinically relevant information faster than traditional culture methods, allowing clinicians to make more efficient and educated antibiotic regimen decisions than with previous methods.5
It takes 24 to 48 hours from blood collection for culture incubation, positivity, and gram staining to occur at VHI. If the gram stain is positive, the blood culture is placed on the BioFire BCID2 in addition to traditional culture medium. BioFire BCID2 results are ready in 45 to 60 minutes. Results are uploaded into the electronic health record (EHR) ≤ 2 hours after they are obtained and the primary team is notified if the test is positive for certain critical results. Susceptibility testing of an identified organism typically requires an additional 24 to 48 hours for finalization. VHI Infectious Disease created an evidence-based antibiotic recommendation chart for certain medication(s) and alternate therapies based on the reported organism and its interpreted presence of resistance markers (eg, ceftriaxone for Escherichia coli when extended-spectrum beta lactamases are not detected vs meropenem if extended-spectrum beta lactamases marker are present). These charts optimize the antibiotic regimen while awaiting susceptibility finalizations.
Two previous studies describe the impact of rapid diagnostic testing technology at US Department of Veterans Affairs (VA) medical centers.6,7 In Texas, the ASP reviewed BCID panel results via clinical decision support software for about 1 hour per day.6 A Los Angeles study analyzed the impact of Biofire BCID with an interpretation guide centered on unnecessary vancomycin use and determined that shorter duration of the medication may have been the result of more frequent infectious disease consultation.7
This study assessed the time to optimal antibiotic de-escalation before and after the implementation of BioFire BCID2 with results reviewed by the ASP without active notification or assistance of any clinical decision support technology. The primary objective was to evaluate difference in time to optimal antibiotics from blood culture draw pre- vs postintervention. Secondary objectives included differences in time to organism identification, difference in time on broad-spectrum antibiotics, and difference in time to appropriate antibiotics.
Methods
This quasi-experimental retrospective chart review assessed the impact of BioFire BCID2 use on timely antibiotic de-escalation for patients who experienced a BSI at VHI between March 1, 2022, and October 1, 2023. Microbiology laboratory records identified eligible patients with positive blood cultures within the study time frame. Data were collected from the VHI EHR.
Patients were included if they had a positive bacterial blood culture and received ≥ 1 antibiotic indicated for bacteremia while receiving inpatient care. Patients were excluded if they died prior to blood culture results, transferred out of VHI, left against medical advice, or had untreated contaminants in blood culture results (ie, never received antibiotics aimed at the contaminated culture).
Patient lists were generated for before and after implementation of BioFire BCID2 (pre- and postintervention) using the VHI EHR and microbiology laboratory record system. The pre- and postinterventions groups were different sizes. As a result, a random sampling of the preintervention group was selected and included patients from March 1, 2022, through March 26, 2023. The postintervention group was smaller due to time constraints between initiation of BioFire BCID2 for data collection and included all patients from March 27, 2023, through October 1, 2023.
Optimal antibiotics were defined as escalation from inappropriate therapy to broader agent(s), de-escalation from broad-spectrum therapy to targeted agent(s), discontinuation of therapy due to an organism being identified as a contaminant, or optimization of a regimen to the preferred antimicrobial agent based on evidence-based consensus guidelines. Broad-spectrum antibiotics included: piperacillin/tazobactam, cefepime, ceftazidime, ceftazidime-avibactam, cefiderocol, carbapenems, fluroquinolones, vancomycin, daptomycin, ceftaroline, linezolid, or aztreonam. Appropriate antibiotics were defined as those with activity toward the final identified organism(s).
Deidentified participant data were entered into Microsoft Excel and kept on a secure VA server to complete statistical analyses. Parametric continuous data, such as age, were analyzed using the t-test, while nonparametric continuous data, such as time to optimal antibiotics, were analyzed using the Mann-Whitney U test. Categorical data, like sex and race, were analyzed using either Fisher exact test for small sample sizes or X2 test for a larger sample size. Statistical significance levels was defined as P < .05.
Results
Using patient lists drawn from the EHR and the microbiology laboratory records, 110 electronic charts were randomly selected for review. Fifteen patients were excluded: 8 had untreated contaminants, 4 died, and 3 were transferred out of VHI. Of the 95 patients included, 48 were in the preintervention group and 47 were in the postintervention group (Figure 1).

Baseline characteristics were similar between the 2 groups (Table 1). Most patients were White males aged > 70 years in the EHR. The urinary tract was the most common source of infection, impacting 12 patients in each group (Figure 2). Escherichia coli, Klebsiella, Staphylococcus, and Streptococcus were the most common bloodstream isolates identified.


The median time to optimal antibiotics in the preintervention group was 58.5 hours vs 43.4 hours in the postintervention group (P = .11). The median time to organism identification was 37.8 hours in the preintervention group vs 16.9 hours in the postintervention group (P < .001). The median time on broad-spectrum antibiotics was 45.2 hours in the preintervention group vs 46.6 hours in the postintervention group (P = .99). The median time on appropriate antibiotics in the preintervention group was 2.3 hours vs 1.9 hours in the postintervention group (P = .79). Differences in other measured outcomes between the groups were not statistically significant (Table 2).

Although implementation of rapid diagnostic technology reduced the median time to optimal antibiotics, the results were not statistically significant. Shorter time to organism identification in the postintervention group compared to the preintervention group was the lone statistically significant metric (P < .001).
Discussion
A lack of statistical significance in the primary outcome may have been due to nonadherence to facility de-escalation protocols or a suboptimal BioFire BCID2 result notification system. Additionally, use of rapid BCID at VHI may improve over time as clinicians become more familiar with the technology. Gaps in clinical pharmacy coverage during the night shift may have also contributed to delays in antibiotic optimization, particularly if other clinicians are not equipped with the knowledge or training to appropriately deescalate antibiotics based on microorganisms identified. A 2017 study by Donner et al concluded that physician interpretation of BCID results is suboptimal and should be augmented with clinical decision support tools as new technology becomes available.8 Despite the statistically insignificant results of this study, it did highlight potential areas of improvement which can lead to improved patient care.
Previous research has evaluated the impact of rapid BCID technology on antibiotic treatment and clinical outcomes. Chiasson et al found that median time to optimal therapy was 73.8 hours in the pre-BCID arm compared to 34.7 hours in the post- BCID arm (P ≤ .001), emphasizing the importance of combining rapid BCID with clinical decision support tools and pharmacy input.6 Senok et al found that BCID2 implementation led to a significant decrease in median time to culture result, which informed optimal antibiotic therapy and decreased 30-day mortality in the intensive care setting.9 In contrast, the current study did not stratify patients according to medical ward or illness severity even though clinicians may be less likely to de-escalate antibiotic therapy in critically ill patients.
Bae et al reported findings consistent with the current study and concluded that BCID did not affect the clinical outcomes of overall BSIs; however, it contributed to early administration of effective antibiotics in cases of BSIs caused by multidrug-resistant organisms.10 Results of this study were not stratified according to multidrug-resistant organisms because the sample size was too small. The current study also included patients with polymicrobial infections, which may have impacted the results due to a less streamlined approach to antibiotic optimization.
Limitations
This single-center, retrospective study had a small sample size, short time frame, and lacked patient diversity, and therefore may not be generalizable to other health care systems. The sample size was limited by shorter date range and smaller patient list between BioFire BCID2 implementation and data collection, which was used to determine the number of charts selected in each group. Some patients received antibiotics prior to blood cultures being drawn, which may falsely decrease time to optimal/ appropriate antibiotics and falsely increase time on broad spectrum/any antibiotics due to early antibiotic administration. The total number of patients on broad-spectrum antibiotics differed from the total number of patients for other outcomes because several patients never received the defined broad spectrum antibiotics.
Conclusions
When combined with a pre-existing ASP without active notification, the implementation of BioFire BCID2 did not return statistically significant data showing a decrease in time to optimal antibiotics, time to appropriate antibiotics, or time on broad-spectrum antibiotics at VHI. To make this program more successful, pharmacist intervention and clinical decision support tools may be needed.
Additional research is required to determine the optimal integration of antimicrobial stewardship, rapid diagnostic technology, and pharmacy services for maximum benefit. Even though the primary outcome was not statistically significant, the results may be clinically significant from a stewardship perspective. Realigning microbiology workflows to mimic other research, which emphasizes the importance of funneling rapid BCID results through the ASP, may improve outcomes. Future studies may be warranted following the implementation of clinical decision support tools to assess their impact on stewardship practices and patient outcomes.
- Goto M, Al-Hasan MN. Overall burden of bloodstream infection and nosocomial bloodstream infection in North America and Europe. Clin Microbiol Infect. 2013;19(6):501- 509. doi:10.1111/1469-0691.12195
- Pardo J, Klinker KP, Borgert SJ, Butler BM, Giglio PG, Rand KH. Clinical and economic impact of antimicrobial stewardship interventions with the FilmArray blood culture identification panel. Diagn Microbiol Infect Dis. 2016;84(2):159-164. doi:10.1016/j.diagmicrobio.2015.10.023.
- Barlam TF, Cosgrove SE, Abbo LM, et al. Implementing an antibiotic stewardship program: guidelines by the Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America. Clin Infect Dis. 2016;62(10):e51-e77. doi:10.1093/cid/ciw118
- BIOFIRE® Blood Culture Identification 2 (BCID2) Panel. Biomerierux. Updated 2025. Accessed May 10, 2025. https://www.biofiredx.com/products/the-filmarray-panels/filmarraybcid/
- Huang AM, Newton D, Kunapuli A, et al. Impact of rapid organism identification via matrix-assisted laser desorption/ionization time-of-flight combined with antimicrobial stewardship team intervention in adult patients with bacteremia and candidemia. Clin Infect Dis. 2013;57(9):1237-1245. doi:10.1093/cid/cit498
- Chiasson JM, Smith WJ, Jodlowski TZ, Kouma MA, Cutrell JB. Impact of a rapid blood culture diagnostic panel on time to optimal antimicrobial therapy at a veterans affairs medical center. J Pharm Pract. 2022;35(5):722-729. doi:10.1177/08971900211000686
- Wu S, Watson RL, Graber CJ. 2007. Impact of combining rapid diagnostics with an interpretation guide on vancomycin usage for contaminant blood cultures growing coagulase- negative staphylococci (CoNS). Open Forum Infect Dis. 2019;6(Suppl 2):S674. doi:10.1093/ofid/ofz360.1687
- Donner LM, Campbell WS, Lyden E, Van Schooneveld TC. Assessment of rapid-blood-culture-identification result interpretation and antibiotic prescribing practices. J Clin Microbiol. 2017;55(5):1496-1507. doi:10.1128/JCM.02395-16
- Senok A, Dabal LA, Alfaresi M, et al. Clinical impact of the BIOFIRE blood culture identification 2 panel in adult patients with bloodstream infection: a multicentre observational study in the United Arab Emirates. Diagnostics (Basel). 2023;13(14):2433. doi:10.3390/diagnostics13142433
- Bae JY, Bae J, So MK, Choi HJ, Lee M. The impact of the rapid blood culture identification panel on antibiotic treatment and clinical outcomes in bloodstream infections, particularly those associated with multidrug-resistant micro-organisms. Diagnostics (Basel). 2023;13(23):3504. doi:10.3390/diagnostics13233504
- Goto M, Al-Hasan MN. Overall burden of bloodstream infection and nosocomial bloodstream infection in North America and Europe. Clin Microbiol Infect. 2013;19(6):501- 509. doi:10.1111/1469-0691.12195
- Pardo J, Klinker KP, Borgert SJ, Butler BM, Giglio PG, Rand KH. Clinical and economic impact of antimicrobial stewardship interventions with the FilmArray blood culture identification panel. Diagn Microbiol Infect Dis. 2016;84(2):159-164. doi:10.1016/j.diagmicrobio.2015.10.023.
- Barlam TF, Cosgrove SE, Abbo LM, et al. Implementing an antibiotic stewardship program: guidelines by the Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America. Clin Infect Dis. 2016;62(10):e51-e77. doi:10.1093/cid/ciw118
- BIOFIRE® Blood Culture Identification 2 (BCID2) Panel. Biomerierux. Updated 2025. Accessed May 10, 2025. https://www.biofiredx.com/products/the-filmarray-panels/filmarraybcid/
- Huang AM, Newton D, Kunapuli A, et al. Impact of rapid organism identification via matrix-assisted laser desorption/ionization time-of-flight combined with antimicrobial stewardship team intervention in adult patients with bacteremia and candidemia. Clin Infect Dis. 2013;57(9):1237-1245. doi:10.1093/cid/cit498
- Chiasson JM, Smith WJ, Jodlowski TZ, Kouma MA, Cutrell JB. Impact of a rapid blood culture diagnostic panel on time to optimal antimicrobial therapy at a veterans affairs medical center. J Pharm Pract. 2022;35(5):722-729. doi:10.1177/08971900211000686
- Wu S, Watson RL, Graber CJ. 2007. Impact of combining rapid diagnostics with an interpretation guide on vancomycin usage for contaminant blood cultures growing coagulase- negative staphylococci (CoNS). Open Forum Infect Dis. 2019;6(Suppl 2):S674. doi:10.1093/ofid/ofz360.1687
- Donner LM, Campbell WS, Lyden E, Van Schooneveld TC. Assessment of rapid-blood-culture-identification result interpretation and antibiotic prescribing practices. J Clin Microbiol. 2017;55(5):1496-1507. doi:10.1128/JCM.02395-16
- Senok A, Dabal LA, Alfaresi M, et al. Clinical impact of the BIOFIRE blood culture identification 2 panel in adult patients with bloodstream infection: a multicentre observational study in the United Arab Emirates. Diagnostics (Basel). 2023;13(14):2433. doi:10.3390/diagnostics13142433
- Bae JY, Bae J, So MK, Choi HJ, Lee M. The impact of the rapid blood culture identification panel on antibiotic treatment and clinical outcomes in bloodstream infections, particularly those associated with multidrug-resistant micro-organisms. Diagnostics (Basel). 2023;13(23):3504. doi:10.3390/diagnostics13233504
Impact of Rapid Blood Culture Identification on Antibiotic De-escalation at a Veterans Affairs Medical Center
Impact of Rapid Blood Culture Identification on Antibiotic De-escalation at a Veterans Affairs Medical Center
