User login
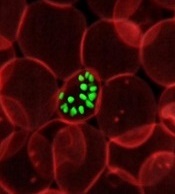
How a malaria parasite is evading treatment
New research has revealed mutations that help the malaria parasite Plasmodium falciparum evade treatment.
Researchers used whole-genome analyses and chemogenetics to identify drug targets and resistance genes in cell lines of P falciparum that are resistant to antimalarial compounds.
The group’s work confirmed previously known mutations that contribute to the parasite’s resistance but also revealed new targets that may deepen our understanding of the parasite’s underlying biology.
“This exploration of the P falciparum resistome—the collection of antibiotic resistance genes—and its druggable genome will help guide new drug discovery efforts and advance our understanding of how the malaria parasite evolves to fight back,” said Elizabeth Winzeler, PhD, of the University of California San Diego School of Medicine.
She and her colleagues conducted this research and reported the results in Science.
“A single human [malaria] infection can result in a person containing upwards of a trillion asexual blood-stage parasites,” Dr Winzeler said. “Even with a relatively slow random mutation rate, these numbers confer extraordinary adaptability.”
“In just a few cycles of replication, the P falciparum genome can acquire a random genetic change that may render at least one parasite resistant to the activity of a drug or human-encoded antibody.”
Such rapid evolution can be exploited in vitro to document how the parasite evolves in the presence of antimalarials, and it can be used to reveal new drug targets.
With this in mind, Dr Winzeler and her colleagues performed a genome analysis of 262 P falciparum parasites resistant to 37 groups of compounds.
In 83 genes associated with drug resistance, the researchers identified hundreds of changes that could be mediating the resistance, including 159 gene amplifications and 148 nonsynonymous mutations.
The team then used clones of well-studied P falciparum parasites and exposed them to the compounds over time to induce resistance, monitoring the genetic changes that occurred as resistance developed.
The researchers were able to identify a likely target or resistance gene for every compound.
In addition, the team identified mutations that repeatedly occurred upon individual exposure to a variety of drugs, meaning these mutations are likely mediating resistance to numerous existing treatments.
“Our findings showed and underscored the challenging complexity of evolved drug resistance in P falciparum, but they also identified new drug targets or resistance genes for every compound for which resistant parasites were generated,” Dr Winzeler said.
“It revealed the complicated chemogenetic landscape of P falciparum but also provided a potential guide for designing new small-molecule inhibitors to fight this pathogen.” ![]()
New research has revealed mutations that help the malaria parasite Plasmodium falciparum evade treatment.
Researchers used whole-genome analyses and chemogenetics to identify drug targets and resistance genes in cell lines of P falciparum that are resistant to antimalarial compounds.
The group’s work confirmed previously known mutations that contribute to the parasite’s resistance but also revealed new targets that may deepen our understanding of the parasite’s underlying biology.
“This exploration of the P falciparum resistome—the collection of antibiotic resistance genes—and its druggable genome will help guide new drug discovery efforts and advance our understanding of how the malaria parasite evolves to fight back,” said Elizabeth Winzeler, PhD, of the University of California San Diego School of Medicine.
She and her colleagues conducted this research and reported the results in Science.
“A single human [malaria] infection can result in a person containing upwards of a trillion asexual blood-stage parasites,” Dr Winzeler said. “Even with a relatively slow random mutation rate, these numbers confer extraordinary adaptability.”
“In just a few cycles of replication, the P falciparum genome can acquire a random genetic change that may render at least one parasite resistant to the activity of a drug or human-encoded antibody.”
Such rapid evolution can be exploited in vitro to document how the parasite evolves in the presence of antimalarials, and it can be used to reveal new drug targets.
With this in mind, Dr Winzeler and her colleagues performed a genome analysis of 262 P falciparum parasites resistant to 37 groups of compounds.
In 83 genes associated with drug resistance, the researchers identified hundreds of changes that could be mediating the resistance, including 159 gene amplifications and 148 nonsynonymous mutations.
The team then used clones of well-studied P falciparum parasites and exposed them to the compounds over time to induce resistance, monitoring the genetic changes that occurred as resistance developed.
The researchers were able to identify a likely target or resistance gene for every compound.
In addition, the team identified mutations that repeatedly occurred upon individual exposure to a variety of drugs, meaning these mutations are likely mediating resistance to numerous existing treatments.
“Our findings showed and underscored the challenging complexity of evolved drug resistance in P falciparum, but they also identified new drug targets or resistance genes for every compound for which resistant parasites were generated,” Dr Winzeler said.
“It revealed the complicated chemogenetic landscape of P falciparum but also provided a potential guide for designing new small-molecule inhibitors to fight this pathogen.” ![]()
New research has revealed mutations that help the malaria parasite Plasmodium falciparum evade treatment.
Researchers used whole-genome analyses and chemogenetics to identify drug targets and resistance genes in cell lines of P falciparum that are resistant to antimalarial compounds.
The group’s work confirmed previously known mutations that contribute to the parasite’s resistance but also revealed new targets that may deepen our understanding of the parasite’s underlying biology.
“This exploration of the P falciparum resistome—the collection of antibiotic resistance genes—and its druggable genome will help guide new drug discovery efforts and advance our understanding of how the malaria parasite evolves to fight back,” said Elizabeth Winzeler, PhD, of the University of California San Diego School of Medicine.
She and her colleagues conducted this research and reported the results in Science.
“A single human [malaria] infection can result in a person containing upwards of a trillion asexual blood-stage parasites,” Dr Winzeler said. “Even with a relatively slow random mutation rate, these numbers confer extraordinary adaptability.”
“In just a few cycles of replication, the P falciparum genome can acquire a random genetic change that may render at least one parasite resistant to the activity of a drug or human-encoded antibody.”
Such rapid evolution can be exploited in vitro to document how the parasite evolves in the presence of antimalarials, and it can be used to reveal new drug targets.
With this in mind, Dr Winzeler and her colleagues performed a genome analysis of 262 P falciparum parasites resistant to 37 groups of compounds.
In 83 genes associated with drug resistance, the researchers identified hundreds of changes that could be mediating the resistance, including 159 gene amplifications and 148 nonsynonymous mutations.
The team then used clones of well-studied P falciparum parasites and exposed them to the compounds over time to induce resistance, monitoring the genetic changes that occurred as resistance developed.
The researchers were able to identify a likely target or resistance gene for every compound.
In addition, the team identified mutations that repeatedly occurred upon individual exposure to a variety of drugs, meaning these mutations are likely mediating resistance to numerous existing treatments.
“Our findings showed and underscored the challenging complexity of evolved drug resistance in P falciparum, but they also identified new drug targets or resistance genes for every compound for which resistant parasites were generated,” Dr Winzeler said.
“It revealed the complicated chemogenetic landscape of P falciparum but also provided a potential guide for designing new small-molecule inhibitors to fight this pathogen.” ![]()
Emicizumab still available despite legal issues
The Roche Group has issued a statement reassuring the US hemophilia community that legal issues are not affecting patient access to emicizumab (Hemlibra), at least for the time being.
Emicizumab is a bispecific factor IXa- and factor X-directed antibody approved in the US as routine prophylaxis to prevent or reduce the frequency of bleeding episodes in adults and children who have hemophilia A and factor VIII (FVIII) inhibitors.
The Roche Group—which consists of Genentech in the US, Chugai in Japan, and Roche in the rest of the world—said emicizumab is still available for these patients, despite a legal battle with Baxalta, a wholly owned subsidiary of Shire.
In May 2017, Baxalta sued Genentech and Chugai for allegedly infringing upon US Patent No. 7,033,590.
The patent covers factor IX/factor IXa antibodies and antibody derivatives. It was issued to Baxter in April 2006 and assigned to Baxalta in March 2016. The patent is still active.
According to Baxalta, Genentech and Chugai are infringing on the patent by manufacturing, selling, or importing emicizumab. Therefore, Baxalta is seeking judgment in its favor and monetary damages.
The trial for this case is scheduled for September 2019.
However, in December 2017, Baxalta filed a motion for a preliminary injunction that would prevent the sale of emicizumab in the US. If granted, the injunction would prevent the following patients from receiving emicizumab:
- Hemophilia A patients with inhibitors (an inhibitor titer of greater than 5 Bethesda units) who cannot be treated effectively with FVIII replacement therapy, unless (i) they have already started emicizumab before the injunction is granted or (ii) they have previous experience with on-demand or prophylactic bypassing agents and their needs are not being met, as defined by Shire using criteria that include experiencing certain life- or limb-threatening bleeds or venous access issues.
- Hemophilia A patients who have an inhibitor titer less than or equal to 5 Bethesda units or who can be effectively treated with FVIII replacement therapy, regardless of whether they have already started emicizumab.
- Hemophilia A patients without inhibitors, regardless of whether they have already started emicizumab.
The Roche Group said it believes Baxalta’s claim is not valid, emicizumab does not infringe upon the patent, and the group will oppose the injunction.
The court’s decision on the injunction is expected this summer. In the meantime, emicizumab is still available for the aforementioned patients. ![]()
The Roche Group has issued a statement reassuring the US hemophilia community that legal issues are not affecting patient access to emicizumab (Hemlibra), at least for the time being.
Emicizumab is a bispecific factor IXa- and factor X-directed antibody approved in the US as routine prophylaxis to prevent or reduce the frequency of bleeding episodes in adults and children who have hemophilia A and factor VIII (FVIII) inhibitors.
The Roche Group—which consists of Genentech in the US, Chugai in Japan, and Roche in the rest of the world—said emicizumab is still available for these patients, despite a legal battle with Baxalta, a wholly owned subsidiary of Shire.
In May 2017, Baxalta sued Genentech and Chugai for allegedly infringing upon US Patent No. 7,033,590.
The patent covers factor IX/factor IXa antibodies and antibody derivatives. It was issued to Baxter in April 2006 and assigned to Baxalta in March 2016. The patent is still active.
According to Baxalta, Genentech and Chugai are infringing on the patent by manufacturing, selling, or importing emicizumab. Therefore, Baxalta is seeking judgment in its favor and monetary damages.
The trial for this case is scheduled for September 2019.
However, in December 2017, Baxalta filed a motion for a preliminary injunction that would prevent the sale of emicizumab in the US. If granted, the injunction would prevent the following patients from receiving emicizumab:
- Hemophilia A patients with inhibitors (an inhibitor titer of greater than 5 Bethesda units) who cannot be treated effectively with FVIII replacement therapy, unless (i) they have already started emicizumab before the injunction is granted or (ii) they have previous experience with on-demand or prophylactic bypassing agents and their needs are not being met, as defined by Shire using criteria that include experiencing certain life- or limb-threatening bleeds or venous access issues.
- Hemophilia A patients who have an inhibitor titer less than or equal to 5 Bethesda units or who can be effectively treated with FVIII replacement therapy, regardless of whether they have already started emicizumab.
- Hemophilia A patients without inhibitors, regardless of whether they have already started emicizumab.
The Roche Group said it believes Baxalta’s claim is not valid, emicizumab does not infringe upon the patent, and the group will oppose the injunction.
The court’s decision on the injunction is expected this summer. In the meantime, emicizumab is still available for the aforementioned patients. ![]()
The Roche Group has issued a statement reassuring the US hemophilia community that legal issues are not affecting patient access to emicizumab (Hemlibra), at least for the time being.
Emicizumab is a bispecific factor IXa- and factor X-directed antibody approved in the US as routine prophylaxis to prevent or reduce the frequency of bleeding episodes in adults and children who have hemophilia A and factor VIII (FVIII) inhibitors.
The Roche Group—which consists of Genentech in the US, Chugai in Japan, and Roche in the rest of the world—said emicizumab is still available for these patients, despite a legal battle with Baxalta, a wholly owned subsidiary of Shire.
In May 2017, Baxalta sued Genentech and Chugai for allegedly infringing upon US Patent No. 7,033,590.
The patent covers factor IX/factor IXa antibodies and antibody derivatives. It was issued to Baxter in April 2006 and assigned to Baxalta in March 2016. The patent is still active.
According to Baxalta, Genentech and Chugai are infringing on the patent by manufacturing, selling, or importing emicizumab. Therefore, Baxalta is seeking judgment in its favor and monetary damages.
The trial for this case is scheduled for September 2019.
However, in December 2017, Baxalta filed a motion for a preliminary injunction that would prevent the sale of emicizumab in the US. If granted, the injunction would prevent the following patients from receiving emicizumab:
- Hemophilia A patients with inhibitors (an inhibitor titer of greater than 5 Bethesda units) who cannot be treated effectively with FVIII replacement therapy, unless (i) they have already started emicizumab before the injunction is granted or (ii) they have previous experience with on-demand or prophylactic bypassing agents and their needs are not being met, as defined by Shire using criteria that include experiencing certain life- or limb-threatening bleeds or venous access issues.
- Hemophilia A patients who have an inhibitor titer less than or equal to 5 Bethesda units or who can be effectively treated with FVIII replacement therapy, regardless of whether they have already started emicizumab.
- Hemophilia A patients without inhibitors, regardless of whether they have already started emicizumab.
The Roche Group said it believes Baxalta’s claim is not valid, emicizumab does not infringe upon the patent, and the group will oppose the injunction.
The court’s decision on the injunction is expected this summer. In the meantime, emicizumab is still available for the aforementioned patients. ![]()
Generic bortezomib available in US
Fresenius Kabi has introduced its generic version of Velcade, Bortezomib for Injection, to the US market.
This is the first intravenous alternative to Velcade available in the US.
Bortezomib for Injection is available as a single dose vial containing 3.5 mg of lyophilized powder.
The product is approved to treat patients with multiple myeloma and patients with mantle cell lymphoma who have received at least 1 prior therapy.
For details, see the prescribing information for Bortezomib for Injection.
Velcade is a registered trademark of Millennium Pharmaceuticals, Inc. ![]()
Fresenius Kabi has introduced its generic version of Velcade, Bortezomib for Injection, to the US market.
This is the first intravenous alternative to Velcade available in the US.
Bortezomib for Injection is available as a single dose vial containing 3.5 mg of lyophilized powder.
The product is approved to treat patients with multiple myeloma and patients with mantle cell lymphoma who have received at least 1 prior therapy.
For details, see the prescribing information for Bortezomib for Injection.
Velcade is a registered trademark of Millennium Pharmaceuticals, Inc. ![]()
Fresenius Kabi has introduced its generic version of Velcade, Bortezomib for Injection, to the US market.
This is the first intravenous alternative to Velcade available in the US.
Bortezomib for Injection is available as a single dose vial containing 3.5 mg of lyophilized powder.
The product is approved to treat patients with multiple myeloma and patients with mantle cell lymphoma who have received at least 1 prior therapy.
For details, see the prescribing information for Bortezomib for Injection.
Velcade is a registered trademark of Millennium Pharmaceuticals, Inc. ![]()
Interventions can increase cord blood donations
Simple interventions can increase cord blood donations, according to research published in Scientific Reports.
Researchers saw a significant increase in cord blood donation when expectant mothers received information about the procedure and were asked to indicate their interest in donating at both early and late stages of their pregnancies.
“We more than doubled the number of cord blood units that were collected,” said study author Nicola Lacetera, PhD, of the University of Toronto Mississauga in Ontario, Canada.
“We learned a lot, and we did a little bit of good too, so that feels nice.”
Dr Lacetera and his colleagues conducted this study in Milan, Italy, where private cord blood banking is banned.
The team set out to determine if providing expectant mothers with information about cord blood donation and prompting them to consider the procedure would increase donations to a public cord blood bank.
Interventions
The researchers enrolled 850 expectant mothers and divided them into 6 treatment cohorts.
The T0 cohort included 217 control subjects who did not receive any information on cord blood donation.
The T1 cohort included 64 subjects who received information on cord blood donation during their first trimester.
The T2 cohort included 88 subjects who were given information on cord blood donation and asked about their intentions to donate in their first trimester.
The T3 cohort included 197 subjects who received information on cord blood donation in their third trimester.
The T4 cohort included 249 subjects who were given information on cord blood donation and asked about their intentions to donate during their third trimester.
The T5 cohort included 35 subjects who were given information on cord blood donation and asked about their intentions to donate during the first trimester and the third trimester.
Results
The researchers found that T5 subjects had the highest donation rate.
In the entire study sample, the donation rate was 2.3% (5/217) in controls, 6.3% (4/64) in T1 subjects, 1.1% (1/88) in T2, 8.1% (16/197) in T3, 10.0% (25/249) in T4, and 17.1% in T5 (6/35).
These results may not be entirely accurate, however, because the researchers could only confirm patients’ donation status if mothers delivered their babies at the study hospital, Ospedale dei Bambini Vittore Buzzi (also known as Buzzi Hospital, BH).
Among women who delivered at BH, donation rates were 2.7% (5/183) in controls, 11.7% (4/34) in T1, 2.2% (1/45) in T2, 8.9% (16/179) in T3, 11.4% (25/42) in T4, and 21.4% (6/28) in T5.
Though these data suggest the various interventions tested can increase cord blood donations, donation rates in this study could have been even higher, according to the researchers.
There were 197 women who submitted consent forms to donate cord blood, were medically eligible to donate, and delivered their babies at BH. However, only 57 of these women successfully donated cord blood.
There were 62 women (56.9%) who could not donate because of medical complications during delivery.
Thirty-three women (30.3%) failed to donate because of organizational reasons, including overcrowding of the delivery room and the absence of obstetric nurses certified to collect and process cord blood at the time of delivery.
There were 14 women (7.1%) who did not donate for institution-related reasons. For example, the women gave birth when the Milan Cord Blood Bank was closed.
There were no details on the remaining 31 women who failed to donate. ![]()
Simple interventions can increase cord blood donations, according to research published in Scientific Reports.
Researchers saw a significant increase in cord blood donation when expectant mothers received information about the procedure and were asked to indicate their interest in donating at both early and late stages of their pregnancies.
“We more than doubled the number of cord blood units that were collected,” said study author Nicola Lacetera, PhD, of the University of Toronto Mississauga in Ontario, Canada.
“We learned a lot, and we did a little bit of good too, so that feels nice.”
Dr Lacetera and his colleagues conducted this study in Milan, Italy, where private cord blood banking is banned.
The team set out to determine if providing expectant mothers with information about cord blood donation and prompting them to consider the procedure would increase donations to a public cord blood bank.
Interventions
The researchers enrolled 850 expectant mothers and divided them into 6 treatment cohorts.
The T0 cohort included 217 control subjects who did not receive any information on cord blood donation.
The T1 cohort included 64 subjects who received information on cord blood donation during their first trimester.
The T2 cohort included 88 subjects who were given information on cord blood donation and asked about their intentions to donate in their first trimester.
The T3 cohort included 197 subjects who received information on cord blood donation in their third trimester.
The T4 cohort included 249 subjects who were given information on cord blood donation and asked about their intentions to donate during their third trimester.
The T5 cohort included 35 subjects who were given information on cord blood donation and asked about their intentions to donate during the first trimester and the third trimester.
Results
The researchers found that T5 subjects had the highest donation rate.
In the entire study sample, the donation rate was 2.3% (5/217) in controls, 6.3% (4/64) in T1 subjects, 1.1% (1/88) in T2, 8.1% (16/197) in T3, 10.0% (25/249) in T4, and 17.1% in T5 (6/35).
These results may not be entirely accurate, however, because the researchers could only confirm patients’ donation status if mothers delivered their babies at the study hospital, Ospedale dei Bambini Vittore Buzzi (also known as Buzzi Hospital, BH).
Among women who delivered at BH, donation rates were 2.7% (5/183) in controls, 11.7% (4/34) in T1, 2.2% (1/45) in T2, 8.9% (16/179) in T3, 11.4% (25/42) in T4, and 21.4% (6/28) in T5.
Though these data suggest the various interventions tested can increase cord blood donations, donation rates in this study could have been even higher, according to the researchers.
There were 197 women who submitted consent forms to donate cord blood, were medically eligible to donate, and delivered their babies at BH. However, only 57 of these women successfully donated cord blood.
There were 62 women (56.9%) who could not donate because of medical complications during delivery.
Thirty-three women (30.3%) failed to donate because of organizational reasons, including overcrowding of the delivery room and the absence of obstetric nurses certified to collect and process cord blood at the time of delivery.
There were 14 women (7.1%) who did not donate for institution-related reasons. For example, the women gave birth when the Milan Cord Blood Bank was closed.
There were no details on the remaining 31 women who failed to donate. ![]()
Simple interventions can increase cord blood donations, according to research published in Scientific Reports.
Researchers saw a significant increase in cord blood donation when expectant mothers received information about the procedure and were asked to indicate their interest in donating at both early and late stages of their pregnancies.
“We more than doubled the number of cord blood units that were collected,” said study author Nicola Lacetera, PhD, of the University of Toronto Mississauga in Ontario, Canada.
“We learned a lot, and we did a little bit of good too, so that feels nice.”
Dr Lacetera and his colleagues conducted this study in Milan, Italy, where private cord blood banking is banned.
The team set out to determine if providing expectant mothers with information about cord blood donation and prompting them to consider the procedure would increase donations to a public cord blood bank.
Interventions
The researchers enrolled 850 expectant mothers and divided them into 6 treatment cohorts.
The T0 cohort included 217 control subjects who did not receive any information on cord blood donation.
The T1 cohort included 64 subjects who received information on cord blood donation during their first trimester.
The T2 cohort included 88 subjects who were given information on cord blood donation and asked about their intentions to donate in their first trimester.
The T3 cohort included 197 subjects who received information on cord blood donation in their third trimester.
The T4 cohort included 249 subjects who were given information on cord blood donation and asked about their intentions to donate during their third trimester.
The T5 cohort included 35 subjects who were given information on cord blood donation and asked about their intentions to donate during the first trimester and the third trimester.
Results
The researchers found that T5 subjects had the highest donation rate.
In the entire study sample, the donation rate was 2.3% (5/217) in controls, 6.3% (4/64) in T1 subjects, 1.1% (1/88) in T2, 8.1% (16/197) in T3, 10.0% (25/249) in T4, and 17.1% in T5 (6/35).
These results may not be entirely accurate, however, because the researchers could only confirm patients’ donation status if mothers delivered their babies at the study hospital, Ospedale dei Bambini Vittore Buzzi (also known as Buzzi Hospital, BH).
Among women who delivered at BH, donation rates were 2.7% (5/183) in controls, 11.7% (4/34) in T1, 2.2% (1/45) in T2, 8.9% (16/179) in T3, 11.4% (25/42) in T4, and 21.4% (6/28) in T5.
Though these data suggest the various interventions tested can increase cord blood donations, donation rates in this study could have been even higher, according to the researchers.
There were 197 women who submitted consent forms to donate cord blood, were medically eligible to donate, and delivered their babies at BH. However, only 57 of these women successfully donated cord blood.
There were 62 women (56.9%) who could not donate because of medical complications during delivery.
Thirty-three women (30.3%) failed to donate because of organizational reasons, including overcrowding of the delivery room and the absence of obstetric nurses certified to collect and process cord blood at the time of delivery.
There were 14 women (7.1%) who did not donate for institution-related reasons. For example, the women gave birth when the Milan Cord Blood Bank was closed.
There were no details on the remaining 31 women who failed to donate. ![]()
EMA recommends orphan designation for pracinostat
The European Medicines Agency (EMA) has recommended that pracinostat receive orphan drug designation.
Pracinostat is an oral histone deacetylase inhibitor currently under investigation in a phase 3 study in combination with azacitidine for the treatment of acute myeloid leukemia (AML) in adult patients unfit to receive induction chemotherapy.
Orphan designation provides regulatory and financial incentives for companies to develop and market therapies that treat life-threatening or chronically debilitating conditions affecting no more than 5 in 10,000 people in the European Union, and where no satisfactory treatment is available.
Orphan designation provides incentives for companies seeking protocol assistance from the EMA during the product development phase and direct access to the centralized authorization procedure. The designation also provides a 10-year period of marketing exclusivity if a therapy receives regulatory approval.
The EMA’s Committee for Orphan Medicinal Products adopts an opinion on the granting of orphan drug designation, and that opinion is submitted to the European Commission for a final decision.
Phase 2 study
The EMA’s recommendation that pracinostat receive orphan drug designation is based on results of a phase 2 study, which were presented at the 2016 ASH Annual Meeting.
The study included 50 patients who had a median age of 75 (range, 66-84). Sixty-six percent of patients had de novo AML, and 34% had secondary AML.
The patients received pracinostat at 60 mg orally on days 1, 3, and 5 of each week for 21 days of each 28-day cycle. They received azacitidine at 75 mg/m2 subcutaneously or intravenously on days 1-7 or days 1-5 and 8-9 (per site preference) of each 28-day cycle.
As of October 15, 2016, 90% of patients had discontinued treatment, 42% due to progressive disease, 28% due to adverse events (AEs), 14% due to patient decision, and 6% due to investigator decision.
Fifty-two percent of patients (n=26) achieved the primary endpoint of complete response (CR) plus CR with incomplete count recovery (CRi) plus morphologic leukemia-free state (MLFS).
Forty-two percent of patients had a CR, 4% had a CRi, and 6% achieved MLFS. The median duration of CR/CRi/MLFS was 13.2 months. The median duration of CR/CRi was 17.2 months.
The median overall survival was 19.1 months. The 1-year survival rate was 62%, and the 2-year survival rate was 41%.
The most common treatment-emergent AEs were nausea (78%), constipation (70%), fatigue (62%), decreased appetite (56%), diarrhea (50%), vomiting (40%), cough (36%), dyspnea (34%), hypokalemia (34%), peripheral edema (34%), pyrexia (34%), dizziness (32%), back pain (28%), insomnia (28%), febrile neutropenia (48%), thrombocytopenia (46%), anemia (38%), and neutropenia (38%).
Treatment-emergent AEs led to discontinuation in 14 patients. Three of these patients developed sepsis that proved fatal.
The other AEs leading to discontinuation included grade 3 acute axonal neuropathy, grade 3 parainfluenza, grade 3 prolonged QTc/atrial fibrillation, grade 1 acute kidney injury, grade 3 diverticulitis, grade 3 supraglottic ulcer, grade 2 upper respiratory infection, grade 3 fatigue (n=2), and grades 1 and 3 intermittent fatigue (n=2). ![]()
The European Medicines Agency (EMA) has recommended that pracinostat receive orphan drug designation.
Pracinostat is an oral histone deacetylase inhibitor currently under investigation in a phase 3 study in combination with azacitidine for the treatment of acute myeloid leukemia (AML) in adult patients unfit to receive induction chemotherapy.
Orphan designation provides regulatory and financial incentives for companies to develop and market therapies that treat life-threatening or chronically debilitating conditions affecting no more than 5 in 10,000 people in the European Union, and where no satisfactory treatment is available.
Orphan designation provides incentives for companies seeking protocol assistance from the EMA during the product development phase and direct access to the centralized authorization procedure. The designation also provides a 10-year period of marketing exclusivity if a therapy receives regulatory approval.
The EMA’s Committee for Orphan Medicinal Products adopts an opinion on the granting of orphan drug designation, and that opinion is submitted to the European Commission for a final decision.
Phase 2 study
The EMA’s recommendation that pracinostat receive orphan drug designation is based on results of a phase 2 study, which were presented at the 2016 ASH Annual Meeting.
The study included 50 patients who had a median age of 75 (range, 66-84). Sixty-six percent of patients had de novo AML, and 34% had secondary AML.
The patients received pracinostat at 60 mg orally on days 1, 3, and 5 of each week for 21 days of each 28-day cycle. They received azacitidine at 75 mg/m2 subcutaneously or intravenously on days 1-7 or days 1-5 and 8-9 (per site preference) of each 28-day cycle.
As of October 15, 2016, 90% of patients had discontinued treatment, 42% due to progressive disease, 28% due to adverse events (AEs), 14% due to patient decision, and 6% due to investigator decision.
Fifty-two percent of patients (n=26) achieved the primary endpoint of complete response (CR) plus CR with incomplete count recovery (CRi) plus morphologic leukemia-free state (MLFS).
Forty-two percent of patients had a CR, 4% had a CRi, and 6% achieved MLFS. The median duration of CR/CRi/MLFS was 13.2 months. The median duration of CR/CRi was 17.2 months.
The median overall survival was 19.1 months. The 1-year survival rate was 62%, and the 2-year survival rate was 41%.
The most common treatment-emergent AEs were nausea (78%), constipation (70%), fatigue (62%), decreased appetite (56%), diarrhea (50%), vomiting (40%), cough (36%), dyspnea (34%), hypokalemia (34%), peripheral edema (34%), pyrexia (34%), dizziness (32%), back pain (28%), insomnia (28%), febrile neutropenia (48%), thrombocytopenia (46%), anemia (38%), and neutropenia (38%).
Treatment-emergent AEs led to discontinuation in 14 patients. Three of these patients developed sepsis that proved fatal.
The other AEs leading to discontinuation included grade 3 acute axonal neuropathy, grade 3 parainfluenza, grade 3 prolonged QTc/atrial fibrillation, grade 1 acute kidney injury, grade 3 diverticulitis, grade 3 supraglottic ulcer, grade 2 upper respiratory infection, grade 3 fatigue (n=2), and grades 1 and 3 intermittent fatigue (n=2). ![]()
The European Medicines Agency (EMA) has recommended that pracinostat receive orphan drug designation.
Pracinostat is an oral histone deacetylase inhibitor currently under investigation in a phase 3 study in combination with azacitidine for the treatment of acute myeloid leukemia (AML) in adult patients unfit to receive induction chemotherapy.
Orphan designation provides regulatory and financial incentives for companies to develop and market therapies that treat life-threatening or chronically debilitating conditions affecting no more than 5 in 10,000 people in the European Union, and where no satisfactory treatment is available.
Orphan designation provides incentives for companies seeking protocol assistance from the EMA during the product development phase and direct access to the centralized authorization procedure. The designation also provides a 10-year period of marketing exclusivity if a therapy receives regulatory approval.
The EMA’s Committee for Orphan Medicinal Products adopts an opinion on the granting of orphan drug designation, and that opinion is submitted to the European Commission for a final decision.
Phase 2 study
The EMA’s recommendation that pracinostat receive orphan drug designation is based on results of a phase 2 study, which were presented at the 2016 ASH Annual Meeting.
The study included 50 patients who had a median age of 75 (range, 66-84). Sixty-six percent of patients had de novo AML, and 34% had secondary AML.
The patients received pracinostat at 60 mg orally on days 1, 3, and 5 of each week for 21 days of each 28-day cycle. They received azacitidine at 75 mg/m2 subcutaneously or intravenously on days 1-7 or days 1-5 and 8-9 (per site preference) of each 28-day cycle.
As of October 15, 2016, 90% of patients had discontinued treatment, 42% due to progressive disease, 28% due to adverse events (AEs), 14% due to patient decision, and 6% due to investigator decision.
Fifty-two percent of patients (n=26) achieved the primary endpoint of complete response (CR) plus CR with incomplete count recovery (CRi) plus morphologic leukemia-free state (MLFS).
Forty-two percent of patients had a CR, 4% had a CRi, and 6% achieved MLFS. The median duration of CR/CRi/MLFS was 13.2 months. The median duration of CR/CRi was 17.2 months.
The median overall survival was 19.1 months. The 1-year survival rate was 62%, and the 2-year survival rate was 41%.
The most common treatment-emergent AEs were nausea (78%), constipation (70%), fatigue (62%), decreased appetite (56%), diarrhea (50%), vomiting (40%), cough (36%), dyspnea (34%), hypokalemia (34%), peripheral edema (34%), pyrexia (34%), dizziness (32%), back pain (28%), insomnia (28%), febrile neutropenia (48%), thrombocytopenia (46%), anemia (38%), and neutropenia (38%).
Treatment-emergent AEs led to discontinuation in 14 patients. Three of these patients developed sepsis that proved fatal.
The other AEs leading to discontinuation included grade 3 acute axonal neuropathy, grade 3 parainfluenza, grade 3 prolonged QTc/atrial fibrillation, grade 1 acute kidney injury, grade 3 diverticulitis, grade 3 supraglottic ulcer, grade 2 upper respiratory infection, grade 3 fatigue (n=2), and grades 1 and 3 intermittent fatigue (n=2). ![]()
Survival differences among AYAs with blood cancers
A new report has revealed differences in survival among adolescents and young adults (AYAs) with hematologic malignancies.
The report includes information on AYAs—ages 15 to 39—living in Los Angeles County who were diagnosed with common cancers between 1988 and 2014.
The data showed differences in 5-year survival rates according to sex, race, age, and socioeconomic status (SES).
For example, lymphoma survival rates were lower for males, African Americans (AAs), older AYAs, and patients with low socioeconomic status (SES).
For AYAs with leukemias, there was no survival difference according to sex, but AAs had worse survival than patients of other races. And the impact of age and SES varied according to leukemia type.
“Cancer survival data are poorly understood for 15- to 39-year-olds,” noted Amie Hwang, PhD, of the University of Southern California Keck School of Medicine in Los Angeles.
That is why she and her colleagues created the report, “Cancer in Los Angeles County: Survival Among Adolescents and Young Adults 1988-2014.”
According to the authors, this is the first report to break down cancer survival rates for AYAs into segments on race/ethnicity, sex, age group, SES, and cancer stage.
Survival data for patients with hematologic malignancies were as follows.
Acute lymphoblastic leukemia
There were 1137 cases of acute lymphoblastic leukemia in the AYA population in Los Angeles County during the period studied. This included 752 males and 385 females.
Five-year survival was similar between males (43%) and females (41%).
Younger AYAs had better survival than older AYAs (48% for ages 15-24, 35% for ages 25-34, and 32% for ages 35-39).
Survival was highest among non-Latino whites (NLWs, 56%), followed by Asian/Pacific Islanders (APIs, 52%), patients of other/unknown races (51%), Latino whites (LWs, 38%), and AAs (29%).
Survival declined with SES (55% for high, 42% for middle, and 36% for low SES).
Acute myeloid leukemia
There were 1195 cases of acute myeloid leukemia—641 males and 554 females.
Five-year survival was similar for males (40%) and females (43%) as well as for the different age groups (45% for ages 15-24 vs 40% for the older age groups).
Survival was highest among NLWs (44%), followed by LWs (43%), APIs (40%), other/unknown (33%), and AAs (25%).
Survival declined somewhat with SES (49% for high, 39% for middle, and 41% for low SES).
Chronic myeloid leukemia
There were 655 cases of chronic myeloid leukemia—408 males and 247 females.
Five-year survival was similar for males (70%) and females (71%), but it was slightly higher for older AYAs (69% for ages 15-24, 68% for ages 25-34, and 76% for ages 35-39).
Survival was highest among patients in the “other/unknown” race category (76%), followed by LWs (73%), NLWs/APIs (both 72%), and AAs (57%).
Survival declined somewhat with SES (76% for high, 67% for middle, and 68% for low SES).
Hodgkin lymphoma
There were 2993 AYAs diagnosed with Hodgkin lymphoma—1553 males and 1440 females.
The 5-year survival rate was higher in females (93%) than males (86%) and in younger AYAs (93% for ages 15-24, 89% for ages 25-34, and 85% for ages 35-39).
Survival was highest among patients in the “other/unknown” race category (96%), followed by APIs/NLWs (both 91%), LWs (88%), and AAs (83%).
Survival declined with SES (95% for high, 89% for middle, and 83% for low SES).
And survival was lower for patients with advanced-stage disease (93% localized, 94% regional, and 83% distant).
Non-Hodkgin lymphoma
There were 4485 AYAs diagnosed with non-Hodgkin lymphoma during the study period—3064 males and 1421 females.
The 5-year survival rate was higher in females (75%) than males (46%) and in younger AYAs (69% for ages 15-24, 51% for ages 25-34, and 52% for ages 35-39).
Survival was highest among patients in the “other/unknown” race category (88%), followed by APIs (68%), LWs/NLWs (both 53%), and AAs (50%).
Survival declined with SES (68% for high, 54% for middle, and 45% for low SES).
And survival was lower for patients with advanced-stage disease (61% localized, 66% regional, and 46% distant).
“Adolescents and young adults go to the doctor less often because they have this superhero mentality, like they’re invincible,” said author Dennis Deapen, DrPH, of the University of Southern California Keck School of Medicine.
“Once they do go to a health professional, their cancer diagnosis can be delayed because cancer isn’t the first concern doctors have for this age group. It comes as no surprise that patients diagnosed with late-stage cancer have reduced survival rates.” ![]()
A new report has revealed differences in survival among adolescents and young adults (AYAs) with hematologic malignancies.
The report includes information on AYAs—ages 15 to 39—living in Los Angeles County who were diagnosed with common cancers between 1988 and 2014.
The data showed differences in 5-year survival rates according to sex, race, age, and socioeconomic status (SES).
For example, lymphoma survival rates were lower for males, African Americans (AAs), older AYAs, and patients with low socioeconomic status (SES).
For AYAs with leukemias, there was no survival difference according to sex, but AAs had worse survival than patients of other races. And the impact of age and SES varied according to leukemia type.
“Cancer survival data are poorly understood for 15- to 39-year-olds,” noted Amie Hwang, PhD, of the University of Southern California Keck School of Medicine in Los Angeles.
That is why she and her colleagues created the report, “Cancer in Los Angeles County: Survival Among Adolescents and Young Adults 1988-2014.”
According to the authors, this is the first report to break down cancer survival rates for AYAs into segments on race/ethnicity, sex, age group, SES, and cancer stage.
Survival data for patients with hematologic malignancies were as follows.
Acute lymphoblastic leukemia
There were 1137 cases of acute lymphoblastic leukemia in the AYA population in Los Angeles County during the period studied. This included 752 males and 385 females.
Five-year survival was similar between males (43%) and females (41%).
Younger AYAs had better survival than older AYAs (48% for ages 15-24, 35% for ages 25-34, and 32% for ages 35-39).
Survival was highest among non-Latino whites (NLWs, 56%), followed by Asian/Pacific Islanders (APIs, 52%), patients of other/unknown races (51%), Latino whites (LWs, 38%), and AAs (29%).
Survival declined with SES (55% for high, 42% for middle, and 36% for low SES).
Acute myeloid leukemia
There were 1195 cases of acute myeloid leukemia—641 males and 554 females.
Five-year survival was similar for males (40%) and females (43%) as well as for the different age groups (45% for ages 15-24 vs 40% for the older age groups).
Survival was highest among NLWs (44%), followed by LWs (43%), APIs (40%), other/unknown (33%), and AAs (25%).
Survival declined somewhat with SES (49% for high, 39% for middle, and 41% for low SES).
Chronic myeloid leukemia
There were 655 cases of chronic myeloid leukemia—408 males and 247 females.
Five-year survival was similar for males (70%) and females (71%), but it was slightly higher for older AYAs (69% for ages 15-24, 68% for ages 25-34, and 76% for ages 35-39).
Survival was highest among patients in the “other/unknown” race category (76%), followed by LWs (73%), NLWs/APIs (both 72%), and AAs (57%).
Survival declined somewhat with SES (76% for high, 67% for middle, and 68% for low SES).
Hodgkin lymphoma
There were 2993 AYAs diagnosed with Hodgkin lymphoma—1553 males and 1440 females.
The 5-year survival rate was higher in females (93%) than males (86%) and in younger AYAs (93% for ages 15-24, 89% for ages 25-34, and 85% for ages 35-39).
Survival was highest among patients in the “other/unknown” race category (96%), followed by APIs/NLWs (both 91%), LWs (88%), and AAs (83%).
Survival declined with SES (95% for high, 89% for middle, and 83% for low SES).
And survival was lower for patients with advanced-stage disease (93% localized, 94% regional, and 83% distant).
Non-Hodkgin lymphoma
There were 4485 AYAs diagnosed with non-Hodgkin lymphoma during the study period—3064 males and 1421 females.
The 5-year survival rate was higher in females (75%) than males (46%) and in younger AYAs (69% for ages 15-24, 51% for ages 25-34, and 52% for ages 35-39).
Survival was highest among patients in the “other/unknown” race category (88%), followed by APIs (68%), LWs/NLWs (both 53%), and AAs (50%).
Survival declined with SES (68% for high, 54% for middle, and 45% for low SES).
And survival was lower for patients with advanced-stage disease (61% localized, 66% regional, and 46% distant).
“Adolescents and young adults go to the doctor less often because they have this superhero mentality, like they’re invincible,” said author Dennis Deapen, DrPH, of the University of Southern California Keck School of Medicine.
“Once they do go to a health professional, their cancer diagnosis can be delayed because cancer isn’t the first concern doctors have for this age group. It comes as no surprise that patients diagnosed with late-stage cancer have reduced survival rates.” ![]()
A new report has revealed differences in survival among adolescents and young adults (AYAs) with hematologic malignancies.
The report includes information on AYAs—ages 15 to 39—living in Los Angeles County who were diagnosed with common cancers between 1988 and 2014.
The data showed differences in 5-year survival rates according to sex, race, age, and socioeconomic status (SES).
For example, lymphoma survival rates were lower for males, African Americans (AAs), older AYAs, and patients with low socioeconomic status (SES).
For AYAs with leukemias, there was no survival difference according to sex, but AAs had worse survival than patients of other races. And the impact of age and SES varied according to leukemia type.
“Cancer survival data are poorly understood for 15- to 39-year-olds,” noted Amie Hwang, PhD, of the University of Southern California Keck School of Medicine in Los Angeles.
That is why she and her colleagues created the report, “Cancer in Los Angeles County: Survival Among Adolescents and Young Adults 1988-2014.”
According to the authors, this is the first report to break down cancer survival rates for AYAs into segments on race/ethnicity, sex, age group, SES, and cancer stage.
Survival data for patients with hematologic malignancies were as follows.
Acute lymphoblastic leukemia
There were 1137 cases of acute lymphoblastic leukemia in the AYA population in Los Angeles County during the period studied. This included 752 males and 385 females.
Five-year survival was similar between males (43%) and females (41%).
Younger AYAs had better survival than older AYAs (48% for ages 15-24, 35% for ages 25-34, and 32% for ages 35-39).
Survival was highest among non-Latino whites (NLWs, 56%), followed by Asian/Pacific Islanders (APIs, 52%), patients of other/unknown races (51%), Latino whites (LWs, 38%), and AAs (29%).
Survival declined with SES (55% for high, 42% for middle, and 36% for low SES).
Acute myeloid leukemia
There were 1195 cases of acute myeloid leukemia—641 males and 554 females.
Five-year survival was similar for males (40%) and females (43%) as well as for the different age groups (45% for ages 15-24 vs 40% for the older age groups).
Survival was highest among NLWs (44%), followed by LWs (43%), APIs (40%), other/unknown (33%), and AAs (25%).
Survival declined somewhat with SES (49% for high, 39% for middle, and 41% for low SES).
Chronic myeloid leukemia
There were 655 cases of chronic myeloid leukemia—408 males and 247 females.
Five-year survival was similar for males (70%) and females (71%), but it was slightly higher for older AYAs (69% for ages 15-24, 68% for ages 25-34, and 76% for ages 35-39).
Survival was highest among patients in the “other/unknown” race category (76%), followed by LWs (73%), NLWs/APIs (both 72%), and AAs (57%).
Survival declined somewhat with SES (76% for high, 67% for middle, and 68% for low SES).
Hodgkin lymphoma
There were 2993 AYAs diagnosed with Hodgkin lymphoma—1553 males and 1440 females.
The 5-year survival rate was higher in females (93%) than males (86%) and in younger AYAs (93% for ages 15-24, 89% for ages 25-34, and 85% for ages 35-39).
Survival was highest among patients in the “other/unknown” race category (96%), followed by APIs/NLWs (both 91%), LWs (88%), and AAs (83%).
Survival declined with SES (95% for high, 89% for middle, and 83% for low SES).
And survival was lower for patients with advanced-stage disease (93% localized, 94% regional, and 83% distant).
Non-Hodkgin lymphoma
There were 4485 AYAs diagnosed with non-Hodgkin lymphoma during the study period—3064 males and 1421 females.
The 5-year survival rate was higher in females (75%) than males (46%) and in younger AYAs (69% for ages 15-24, 51% for ages 25-34, and 52% for ages 35-39).
Survival was highest among patients in the “other/unknown” race category (88%), followed by APIs (68%), LWs/NLWs (both 53%), and AAs (50%).
Survival declined with SES (68% for high, 54% for middle, and 45% for low SES).
And survival was lower for patients with advanced-stage disease (61% localized, 66% regional, and 46% distant).
“Adolescents and young adults go to the doctor less often because they have this superhero mentality, like they’re invincible,” said author Dennis Deapen, DrPH, of the University of Southern California Keck School of Medicine.
“Once they do go to a health professional, their cancer diagnosis can be delayed because cancer isn’t the first concern doctors have for this age group. It comes as no surprise that patients diagnosed with late-stage cancer have reduced survival rates.” ![]()
Marine animals aid development of cytotoxicity assay
Researchers have looked to deep-sea creatures with the goal of creating a better cytotoxicity assay.
The team harnessed the power of enzymes responsible for marine animal bioluminescence to create the “Matador assay,” which can be used to determine whether cellular and immune-therapeutic agents are actually killing target cells.
The researchers said the Matador assay is quick and simple as well as “highly sensitive,” with the ability to detect cytotoxicity induced by several types of therapies.
Preet M. Chaudhary, MD, PhD, of the University of Southern California Keck School of Medicine in Los Angeles, and his colleagues described the assay in Scientific Reports.
“One of the most promising areas in cancer research is immunotherapy. . .,” Dr Chaudhary said. “It is also one of the most difficult because the methods for testing immunotherapies are not ideal.”
“Radioactive chromium release assay is the gold standard for testing whether an immunotherapy kills cancer cells. This method is expensive, complicated, and requires special disposal practices. Other available methods also suffer from limitations and don’t allow scientists to rapidly screen immunotherapeutic agents to find the best candidates.”
Dr Chaudhary and his colleagues set out to develop a simple, precise, and inexpensive cytotoxicity assay based on marine animal luciferases, the enzymes responsible for bioluminescence.
The team used a group of small crustaceans and deep-sea shrimp, which were selected for their bright bioluminescence. Their luciferases became the basis of the Matador assay.
Engineered to get trapped inside cells, the luciferases leak out of cells when they die, causing a visible glow. The level of luminescence can then be measured with a luminometer.
To test the Matador assay’s effectiveness at measuring cell death, the researchers used several types of cancer cells, including chronic myelogenous leukemia, acute myelogenous leukemia, Burkitt lymphoma, and solid tumor cells.
The team treated these cells with a variety of therapies, including chimeric antigen receptor (CAR) T cells, bispecific T-cell engagers, monoclonal antibodies, and natural killer cells.
Results showed the Matador assay could detect the death of a single cell, a level of sensitivity superior to that of existing cytotoxicity assays.
The researchers also pointed out that the Matador assay is fast, inexpensive, and can be performed in a 384-well plate format, saving time and reagents.
“In our hands, the Matador assay can detect cell death in as little as 30 minutes, which can ultimately translate to more expedient treatments for patients getting cellular immunotherapies such as CAR T cells,” Dr Chaudhary said.
In fact, Dr Chaudhary’s lab has developed more than 75 cancer cell lines expressing the marine luciferases and used them with the Matador assay to develop next-generation CAR T cells.
Dr Chaudhary believes the Matador assay has many potential applications in biomedical research and cellular therapy manufacturing.
“It could potentially play a role in screening other types of anticancer agents or even measuring environmental toxins,” he said. ![]()
Researchers have looked to deep-sea creatures with the goal of creating a better cytotoxicity assay.
The team harnessed the power of enzymes responsible for marine animal bioluminescence to create the “Matador assay,” which can be used to determine whether cellular and immune-therapeutic agents are actually killing target cells.
The researchers said the Matador assay is quick and simple as well as “highly sensitive,” with the ability to detect cytotoxicity induced by several types of therapies.
Preet M. Chaudhary, MD, PhD, of the University of Southern California Keck School of Medicine in Los Angeles, and his colleagues described the assay in Scientific Reports.
“One of the most promising areas in cancer research is immunotherapy. . .,” Dr Chaudhary said. “It is also one of the most difficult because the methods for testing immunotherapies are not ideal.”
“Radioactive chromium release assay is the gold standard for testing whether an immunotherapy kills cancer cells. This method is expensive, complicated, and requires special disposal practices. Other available methods also suffer from limitations and don’t allow scientists to rapidly screen immunotherapeutic agents to find the best candidates.”
Dr Chaudhary and his colleagues set out to develop a simple, precise, and inexpensive cytotoxicity assay based on marine animal luciferases, the enzymes responsible for bioluminescence.
The team used a group of small crustaceans and deep-sea shrimp, which were selected for their bright bioluminescence. Their luciferases became the basis of the Matador assay.
Engineered to get trapped inside cells, the luciferases leak out of cells when they die, causing a visible glow. The level of luminescence can then be measured with a luminometer.
To test the Matador assay’s effectiveness at measuring cell death, the researchers used several types of cancer cells, including chronic myelogenous leukemia, acute myelogenous leukemia, Burkitt lymphoma, and solid tumor cells.
The team treated these cells with a variety of therapies, including chimeric antigen receptor (CAR) T cells, bispecific T-cell engagers, monoclonal antibodies, and natural killer cells.
Results showed the Matador assay could detect the death of a single cell, a level of sensitivity superior to that of existing cytotoxicity assays.
The researchers also pointed out that the Matador assay is fast, inexpensive, and can be performed in a 384-well plate format, saving time and reagents.
“In our hands, the Matador assay can detect cell death in as little as 30 minutes, which can ultimately translate to more expedient treatments for patients getting cellular immunotherapies such as CAR T cells,” Dr Chaudhary said.
In fact, Dr Chaudhary’s lab has developed more than 75 cancer cell lines expressing the marine luciferases and used them with the Matador assay to develop next-generation CAR T cells.
Dr Chaudhary believes the Matador assay has many potential applications in biomedical research and cellular therapy manufacturing.
“It could potentially play a role in screening other types of anticancer agents or even measuring environmental toxins,” he said. ![]()
Researchers have looked to deep-sea creatures with the goal of creating a better cytotoxicity assay.
The team harnessed the power of enzymes responsible for marine animal bioluminescence to create the “Matador assay,” which can be used to determine whether cellular and immune-therapeutic agents are actually killing target cells.
The researchers said the Matador assay is quick and simple as well as “highly sensitive,” with the ability to detect cytotoxicity induced by several types of therapies.
Preet M. Chaudhary, MD, PhD, of the University of Southern California Keck School of Medicine in Los Angeles, and his colleagues described the assay in Scientific Reports.
“One of the most promising areas in cancer research is immunotherapy. . .,” Dr Chaudhary said. “It is also one of the most difficult because the methods for testing immunotherapies are not ideal.”
“Radioactive chromium release assay is the gold standard for testing whether an immunotherapy kills cancer cells. This method is expensive, complicated, and requires special disposal practices. Other available methods also suffer from limitations and don’t allow scientists to rapidly screen immunotherapeutic agents to find the best candidates.”
Dr Chaudhary and his colleagues set out to develop a simple, precise, and inexpensive cytotoxicity assay based on marine animal luciferases, the enzymes responsible for bioluminescence.
The team used a group of small crustaceans and deep-sea shrimp, which were selected for their bright bioluminescence. Their luciferases became the basis of the Matador assay.
Engineered to get trapped inside cells, the luciferases leak out of cells when they die, causing a visible glow. The level of luminescence can then be measured with a luminometer.
To test the Matador assay’s effectiveness at measuring cell death, the researchers used several types of cancer cells, including chronic myelogenous leukemia, acute myelogenous leukemia, Burkitt lymphoma, and solid tumor cells.
The team treated these cells with a variety of therapies, including chimeric antigen receptor (CAR) T cells, bispecific T-cell engagers, monoclonal antibodies, and natural killer cells.
Results showed the Matador assay could detect the death of a single cell, a level of sensitivity superior to that of existing cytotoxicity assays.
The researchers also pointed out that the Matador assay is fast, inexpensive, and can be performed in a 384-well plate format, saving time and reagents.
“In our hands, the Matador assay can detect cell death in as little as 30 minutes, which can ultimately translate to more expedient treatments for patients getting cellular immunotherapies such as CAR T cells,” Dr Chaudhary said.
In fact, Dr Chaudhary’s lab has developed more than 75 cancer cell lines expressing the marine luciferases and used them with the Matador assay to develop next-generation CAR T cells.
Dr Chaudhary believes the Matador assay has many potential applications in biomedical research and cellular therapy manufacturing.
“It could potentially play a role in screening other types of anticancer agents or even measuring environmental toxins,” he said.
DNA methylation may predict outcomes in JMML
New research suggests DNA methylation may be used to predict how children with juvenile myelomonocytic leukemia (JMML) will respond to treatment.
Researchers found they could divide patients into risk groups according to their methylation levels.
Patients with the highest methylation levels had the worst event-free survival, while patients who recovered spontaneously had methylation levels similar to those of healthy control subjects.
“This data provides important information that will help clinicians decide how intensively and swiftly to treat their patients,” said Mignon Loh, MD, of Benioff Children’s Hospital, University of California, San Francisco.
She and her colleagues described this research in Nature Communications.
The team studied genome-wide DNA methylation levels in an initial group of 39 patients with JMML and then validated the results in a group of 40 JMML patients.
Statistical analysis revealed that patients fell into 3 clusters with high, intermediate, or low levels of DNA methylation. And patients’ methylation levels were associated with 4-year event-free survival.
In the initial cohort, 6% (1/15) of patients in the lowest methylation group had an event at 4 years, as did 45% (5/11) of patients with intermediate levels of methylation and 61% (8/13) of patients with the highest levels of methylation.
In the validation cohort, 8% (1/12) of patients in the lowest methylation group had an event at 4 years, as did 36% (4/11) of patients with intermediate levels of methylation and 76% (13/17) of patients with the highest levels of methylation.
“For us, this was surprising,” said study author Elliot Stieglitz, MD, also of Benioff Children’s Hospital.
“We are not yet able to say why DNA methylation is different amongst these patients, but for it to be so predictive of outcomes, even more than genetic mutations, was a big surprise.”
The researchers also found that methylation levels might predict spontaneous remission as well.
Thirteen of the 14 patients who had spontaneous remission clustered together. And their DNA methylation levels were closer to those of healthy control subjects than those of other JMML cases.
“Because we have never been able to tell which patients might spontaneously recover, and because the disease can be so aggressive, the standard of care has been to recommend transplants for everybody,” Dr Stieglitz said.
“There are many health risks associated with bone marrow transplants, ranging from infections to long-term short stature and even death in some cases. To be able to avoid this intensive intervention for some patients based on a methylation assay would really be cutting-edge clinical science.”
New research suggests DNA methylation may be used to predict how children with juvenile myelomonocytic leukemia (JMML) will respond to treatment.
Researchers found they could divide patients into risk groups according to their methylation levels.
Patients with the highest methylation levels had the worst event-free survival, while patients who recovered spontaneously had methylation levels similar to those of healthy control subjects.
“This data provides important information that will help clinicians decide how intensively and swiftly to treat their patients,” said Mignon Loh, MD, of Benioff Children’s Hospital, University of California, San Francisco.
She and her colleagues described this research in Nature Communications.
The team studied genome-wide DNA methylation levels in an initial group of 39 patients with JMML and then validated the results in a group of 40 JMML patients.
Statistical analysis revealed that patients fell into 3 clusters with high, intermediate, or low levels of DNA methylation. And patients’ methylation levels were associated with 4-year event-free survival.
In the initial cohort, 6% (1/15) of patients in the lowest methylation group had an event at 4 years, as did 45% (5/11) of patients with intermediate levels of methylation and 61% (8/13) of patients with the highest levels of methylation.
In the validation cohort, 8% (1/12) of patients in the lowest methylation group had an event at 4 years, as did 36% (4/11) of patients with intermediate levels of methylation and 76% (13/17) of patients with the highest levels of methylation.
“For us, this was surprising,” said study author Elliot Stieglitz, MD, also of Benioff Children’s Hospital.
“We are not yet able to say why DNA methylation is different amongst these patients, but for it to be so predictive of outcomes, even more than genetic mutations, was a big surprise.”
The researchers also found that methylation levels might predict spontaneous remission as well.
Thirteen of the 14 patients who had spontaneous remission clustered together. And their DNA methylation levels were closer to those of healthy control subjects than those of other JMML cases.
“Because we have never been able to tell which patients might spontaneously recover, and because the disease can be so aggressive, the standard of care has been to recommend transplants for everybody,” Dr Stieglitz said.
“There are many health risks associated with bone marrow transplants, ranging from infections to long-term short stature and even death in some cases. To be able to avoid this intensive intervention for some patients based on a methylation assay would really be cutting-edge clinical science.”
New research suggests DNA methylation may be used to predict how children with juvenile myelomonocytic leukemia (JMML) will respond to treatment.
Researchers found they could divide patients into risk groups according to their methylation levels.
Patients with the highest methylation levels had the worst event-free survival, while patients who recovered spontaneously had methylation levels similar to those of healthy control subjects.
“This data provides important information that will help clinicians decide how intensively and swiftly to treat their patients,” said Mignon Loh, MD, of Benioff Children’s Hospital, University of California, San Francisco.
She and her colleagues described this research in Nature Communications.
The team studied genome-wide DNA methylation levels in an initial group of 39 patients with JMML and then validated the results in a group of 40 JMML patients.
Statistical analysis revealed that patients fell into 3 clusters with high, intermediate, or low levels of DNA methylation. And patients’ methylation levels were associated with 4-year event-free survival.
In the initial cohort, 6% (1/15) of patients in the lowest methylation group had an event at 4 years, as did 45% (5/11) of patients with intermediate levels of methylation and 61% (8/13) of patients with the highest levels of methylation.
In the validation cohort, 8% (1/12) of patients in the lowest methylation group had an event at 4 years, as did 36% (4/11) of patients with intermediate levels of methylation and 76% (13/17) of patients with the highest levels of methylation.
“For us, this was surprising,” said study author Elliot Stieglitz, MD, also of Benioff Children’s Hospital.
“We are not yet able to say why DNA methylation is different amongst these patients, but for it to be so predictive of outcomes, even more than genetic mutations, was a big surprise.”
The researchers also found that methylation levels might predict spontaneous remission as well.
Thirteen of the 14 patients who had spontaneous remission clustered together. And their DNA methylation levels were closer to those of healthy control subjects than those of other JMML cases.
“Because we have never been able to tell which patients might spontaneously recover, and because the disease can be so aggressive, the standard of care has been to recommend transplants for everybody,” Dr Stieglitz said.
“There are many health risks associated with bone marrow transplants, ranging from infections to long-term short stature and even death in some cases. To be able to avoid this intensive intervention for some patients based on a methylation assay would really be cutting-edge clinical science.”
Gene variants associated with high-risk pediatric ALL
New research has revealed germline variations associated with high-risk acute lymphoblastic leukemia (ALL) in children.
Researchers sequenced the TP53 tumor suppressor gene in nearly 4000 children with ALL and identified 22 pathogenic germline variants.
These variants were associated with inferior survival and an increased risk of developing second malignancies.
Jun J. Yang, PhD, of St. Jude Children’s Research Hospital in Memphis, Tennessee, and his colleagues reported these findings in the Journal of Clinical Oncology.
The researchers performed targeted sequencing of TP53 coding regions in 3801 children with ALL who were enrolled in 2 Children’s Oncology Group trials (AALL0232 and P9900).
The sequencing revealed 49 unique TP53 coding variants, which were found in 77 children. Twenty-two of the variants were pathogenic, and they were found in 26 children.
The researchers also analyzed data from 60,706 control subjects without ALL and found the 22 pathogenic variants were more likely to be found in the ALL patients than controls. The odds ratio was 5.2 (P<0.001).
Among the ALL patients, those who had the pathogenic variants were significantly older at diagnosis than those without the variants, with median ages of 15.5 years and 7.3 years, respectively (P<0.001).
The pathogenic variants were most common in patients with hypodiploid ALL. About 65% of patients who carried the pathogenic variants had hypodiploid ALL, as did 1.2% of children with wild-type genotype (P<0.001).
The pathogenic variants were associated with inferior event-free survival and overall survival as well. The hazard ratios were 4.2 (P<0.001) and 3.9 (P=0.001), respectively.
And the pathogenic variants were associated with a higher risk of second cancers. The 5-year cumulative incidence of second malignancies was 25.1% among patients with the pathogenic variants and 0.7% among patients without the variants (P<0.001).
“These germline variations are a double whammy for carriers,” Dr Yang said. “Not only is their risk of developing leukemia very high, they are also more likely to relapse or develop a second cancer.”
The association between the pathogenic variants and second cancers has prompted Dr Yang and his colleagues to explore ways to help patients manage their risk.
“Maybe these patients should avoid certain ALL therapies in order to reduce their risk of developing another cancer,” Dr Yang said. “I believe this finding may change treatment and follow-up for these high-risk patients.”
Dr Yang and his colleagues also noted that inherited variations in TP53 are a hallmark of Li-Fraumeni syndrome. And the syndrome might partially explain the high rate of second cancers in ALL patients with the pathogenic TP53 variants.
“The ALL treatment might have added to that risk,” Dr Yang said, “but we do not know for sure.”
New research has revealed germline variations associated with high-risk acute lymphoblastic leukemia (ALL) in children.
Researchers sequenced the TP53 tumor suppressor gene in nearly 4000 children with ALL and identified 22 pathogenic germline variants.
These variants were associated with inferior survival and an increased risk of developing second malignancies.
Jun J. Yang, PhD, of St. Jude Children’s Research Hospital in Memphis, Tennessee, and his colleagues reported these findings in the Journal of Clinical Oncology.
The researchers performed targeted sequencing of TP53 coding regions in 3801 children with ALL who were enrolled in 2 Children’s Oncology Group trials (AALL0232 and P9900).
The sequencing revealed 49 unique TP53 coding variants, which were found in 77 children. Twenty-two of the variants were pathogenic, and they were found in 26 children.
The researchers also analyzed data from 60,706 control subjects without ALL and found the 22 pathogenic variants were more likely to be found in the ALL patients than controls. The odds ratio was 5.2 (P<0.001).
Among the ALL patients, those who had the pathogenic variants were significantly older at diagnosis than those without the variants, with median ages of 15.5 years and 7.3 years, respectively (P<0.001).
The pathogenic variants were most common in patients with hypodiploid ALL. About 65% of patients who carried the pathogenic variants had hypodiploid ALL, as did 1.2% of children with wild-type genotype (P<0.001).
The pathogenic variants were associated with inferior event-free survival and overall survival as well. The hazard ratios were 4.2 (P<0.001) and 3.9 (P=0.001), respectively.
And the pathogenic variants were associated with a higher risk of second cancers. The 5-year cumulative incidence of second malignancies was 25.1% among patients with the pathogenic variants and 0.7% among patients without the variants (P<0.001).
“These germline variations are a double whammy for carriers,” Dr Yang said. “Not only is their risk of developing leukemia very high, they are also more likely to relapse or develop a second cancer.”
The association between the pathogenic variants and second cancers has prompted Dr Yang and his colleagues to explore ways to help patients manage their risk.
“Maybe these patients should avoid certain ALL therapies in order to reduce their risk of developing another cancer,” Dr Yang said. “I believe this finding may change treatment and follow-up for these high-risk patients.”
Dr Yang and his colleagues also noted that inherited variations in TP53 are a hallmark of Li-Fraumeni syndrome. And the syndrome might partially explain the high rate of second cancers in ALL patients with the pathogenic TP53 variants.
“The ALL treatment might have added to that risk,” Dr Yang said, “but we do not know for sure.”
New research has revealed germline variations associated with high-risk acute lymphoblastic leukemia (ALL) in children.
Researchers sequenced the TP53 tumor suppressor gene in nearly 4000 children with ALL and identified 22 pathogenic germline variants.
These variants were associated with inferior survival and an increased risk of developing second malignancies.
Jun J. Yang, PhD, of St. Jude Children’s Research Hospital in Memphis, Tennessee, and his colleagues reported these findings in the Journal of Clinical Oncology.
The researchers performed targeted sequencing of TP53 coding regions in 3801 children with ALL who were enrolled in 2 Children’s Oncology Group trials (AALL0232 and P9900).
The sequencing revealed 49 unique TP53 coding variants, which were found in 77 children. Twenty-two of the variants were pathogenic, and they were found in 26 children.
The researchers also analyzed data from 60,706 control subjects without ALL and found the 22 pathogenic variants were more likely to be found in the ALL patients than controls. The odds ratio was 5.2 (P<0.001).
Among the ALL patients, those who had the pathogenic variants were significantly older at diagnosis than those without the variants, with median ages of 15.5 years and 7.3 years, respectively (P<0.001).
The pathogenic variants were most common in patients with hypodiploid ALL. About 65% of patients who carried the pathogenic variants had hypodiploid ALL, as did 1.2% of children with wild-type genotype (P<0.001).
The pathogenic variants were associated with inferior event-free survival and overall survival as well. The hazard ratios were 4.2 (P<0.001) and 3.9 (P=0.001), respectively.
And the pathogenic variants were associated with a higher risk of second cancers. The 5-year cumulative incidence of second malignancies was 25.1% among patients with the pathogenic variants and 0.7% among patients without the variants (P<0.001).
“These germline variations are a double whammy for carriers,” Dr Yang said. “Not only is their risk of developing leukemia very high, they are also more likely to relapse or develop a second cancer.”
The association between the pathogenic variants and second cancers has prompted Dr Yang and his colleagues to explore ways to help patients manage their risk.
“Maybe these patients should avoid certain ALL therapies in order to reduce their risk of developing another cancer,” Dr Yang said. “I believe this finding may change treatment and follow-up for these high-risk patients.”
Dr Yang and his colleagues also noted that inherited variations in TP53 are a hallmark of Li-Fraumeni syndrome. And the syndrome might partially explain the high rate of second cancers in ALL patients with the pathogenic TP53 variants.
“The ALL treatment might have added to that risk,” Dr Yang said, “but we do not know for sure.”
Method prolongs lifespan of blood samples
A new blood stabilization method significantly prolongs the lifespan of blood samples for microfluidic sorting and transcriptome profiling of rare circulating tumor cells (CTCs), according to researchers.
The method involves reducing the storage temperature to 4° C to reversibly slow down cellular processes while also counteracting the platelet activation that can occur because of the low temperature.
The researchers said this work overcomes a significant barrier to the translation of liquid biopsy technologies for precision oncology and other applications.
Keith Wong, PhD, of the Massachusetts General Hospital Center for Engineering in Medicine (MGH-CEM), and his colleagues described this work in Nature Communications.
When isolating CTCs from fresh, unprocessed blood, timing is everything. Even minor changes in the quality of a blood sample—such as the breakdown of red cells, leukocyte activation, or clot formation— can greatly affect cell-sorting mechanisms and the quality of the biomolecules isolated for cancer detection.
According to published studies, factors such as the total number of CTCs in a sample and the number with high-quality RNA decrease by around 50% within the first 4 to 5 hours after the sample is collected.
“At Mass. General, we have the luxury of being so integrated with the clinical team that we can process blood specimens in the lab typically within an hour or 2 after they are drawn,” Dr Wong said.
“But to make these liquid biopsy technologies routine lab tests for the rest of the world, we need ways to keep blood alive for much longer than several hours, since these assays are best performed in central laboratories for reasons of cost-effectiveness and reproducibility.”
With this in mind, Dr Wong and his colleagues set out to preserve blood in its native state with minimal alterations.
“We wanted to slow down the biological clock as much as possible by using hypothermia, but that is not as simple as it sounds,” said study author Shannon Tessier, PhD, also of MGH-CEM.
“Low temperature is a powerful means to decrease metabolism, but a host of unwanted side effects occur at the same time. In some ways, these challenges are similar to those we face in organ preservation, where we have to optimize strategies for a very complex mix of cells.”
To achieve these goals, the researchers first analyzed the effects of hypothermic storage conditions.
The team found that hypothermic storage (4° C) of blood anticoagulated with acid citrate dextrose maintained “cellular morphology, integrity, and surface epitope stability of diverse hematologic cell types” over 72 hours.
However, the researchers also observed platelet activation.
“We are preserving the blood very well, including the coagulation function of platelets,” Dr Wong said. “But, unfortunately, cooling causes profound activation of platelets. Now, we need a targeted approach for platelets so they don’t form nasty clots in the microfluidic blood-sorting device.”
Fortunately, the researchers found that glycoprotein IIb/IIIa inhibitors were able to counter cooling-induced platelet aggregation. And ion chelation treatment with ethylenediaminetetraacetic acid removed activated platelets from leukocytes.
The team said these steps—hypothermic storage, treatment with glycoprotein IIb/IIIa inhibitors, and ion chelation—allowed whole blood preserved for 3 days to be processed as if it were freshly drawn, with very high purity and virtually no loss in CTC numbers.
“The critical achievement here is that the isolated tumor cells contain high-quality RNA that is suitable for demanding molecular assays, such as single-cell qPCR, droplet digital PCR, and RNA sequencing,” Dr Tessier said.
To test their blood preservation method, Dr Tessier and her colleagues used blood specimens from a group of 10 patients with metastatic prostate cancer.
The researchers compared CTC analysis in preserved blood samples and paired fresh samples from the same patients.
There was 92% agreement in the detection of 12 cancer-specific gene transcripts between the fresh and preserved blood samples.
In addition, there was 100% agreement in the detection of a transcript called AR-V7. Recently published studies showed that the presence of AR-V7 mRNA in prostate cancer CTCs predicts resistance to androgen receptor inhibitors, indicating that chemotherapy may be a better option for such patients.
“The ability to preserve the blood for several days and still be able to pick up this clinically relevant biomarker is remarkable,” said study author David Miyamoto, MD, PhD, of MGH Cancer Center.
“This is very exciting for clinicians because AR-V7 mRNA can only be detected using CTCs and not with circulating tumor DNA or other cell-free assays.”
The researchers highlighted the universal nature of their blood preservation approach by pointing to its compatibility with the microfluidic CTC-iChip device, which isolates tumor cells by rapid removal of blood cells. The team said this suggests the potential impact of this work extends beyond cancer detection.
“With exciting breakthroughs in immunotherapy, stem cell transplantation, and regenerative medicine—in which peripheral blood is often the source of cells for functional assays or ex vivo expansion—the ability to preserve live cells will greatly ease logistical timelines and reduce the cost of complex cell-based assays,” Dr Wong said.
A new blood stabilization method significantly prolongs the lifespan of blood samples for microfluidic sorting and transcriptome profiling of rare circulating tumor cells (CTCs), according to researchers.
The method involves reducing the storage temperature to 4° C to reversibly slow down cellular processes while also counteracting the platelet activation that can occur because of the low temperature.
The researchers said this work overcomes a significant barrier to the translation of liquid biopsy technologies for precision oncology and other applications.
Keith Wong, PhD, of the Massachusetts General Hospital Center for Engineering in Medicine (MGH-CEM), and his colleagues described this work in Nature Communications.
When isolating CTCs from fresh, unprocessed blood, timing is everything. Even minor changes in the quality of a blood sample—such as the breakdown of red cells, leukocyte activation, or clot formation— can greatly affect cell-sorting mechanisms and the quality of the biomolecules isolated for cancer detection.
According to published studies, factors such as the total number of CTCs in a sample and the number with high-quality RNA decrease by around 50% within the first 4 to 5 hours after the sample is collected.
“At Mass. General, we have the luxury of being so integrated with the clinical team that we can process blood specimens in the lab typically within an hour or 2 after they are drawn,” Dr Wong said.
“But to make these liquid biopsy technologies routine lab tests for the rest of the world, we need ways to keep blood alive for much longer than several hours, since these assays are best performed in central laboratories for reasons of cost-effectiveness and reproducibility.”
With this in mind, Dr Wong and his colleagues set out to preserve blood in its native state with minimal alterations.
“We wanted to slow down the biological clock as much as possible by using hypothermia, but that is not as simple as it sounds,” said study author Shannon Tessier, PhD, also of MGH-CEM.
“Low temperature is a powerful means to decrease metabolism, but a host of unwanted side effects occur at the same time. In some ways, these challenges are similar to those we face in organ preservation, where we have to optimize strategies for a very complex mix of cells.”
To achieve these goals, the researchers first analyzed the effects of hypothermic storage conditions.
The team found that hypothermic storage (4° C) of blood anticoagulated with acid citrate dextrose maintained “cellular morphology, integrity, and surface epitope stability of diverse hematologic cell types” over 72 hours.
However, the researchers also observed platelet activation.
“We are preserving the blood very well, including the coagulation function of platelets,” Dr Wong said. “But, unfortunately, cooling causes profound activation of platelets. Now, we need a targeted approach for platelets so they don’t form nasty clots in the microfluidic blood-sorting device.”
Fortunately, the researchers found that glycoprotein IIb/IIIa inhibitors were able to counter cooling-induced platelet aggregation. And ion chelation treatment with ethylenediaminetetraacetic acid removed activated platelets from leukocytes.
The team said these steps—hypothermic storage, treatment with glycoprotein IIb/IIIa inhibitors, and ion chelation—allowed whole blood preserved for 3 days to be processed as if it were freshly drawn, with very high purity and virtually no loss in CTC numbers.
“The critical achievement here is that the isolated tumor cells contain high-quality RNA that is suitable for demanding molecular assays, such as single-cell qPCR, droplet digital PCR, and RNA sequencing,” Dr Tessier said.
To test their blood preservation method, Dr Tessier and her colleagues used blood specimens from a group of 10 patients with metastatic prostate cancer.
The researchers compared CTC analysis in preserved blood samples and paired fresh samples from the same patients.
There was 92% agreement in the detection of 12 cancer-specific gene transcripts between the fresh and preserved blood samples.
In addition, there was 100% agreement in the detection of a transcript called AR-V7. Recently published studies showed that the presence of AR-V7 mRNA in prostate cancer CTCs predicts resistance to androgen receptor inhibitors, indicating that chemotherapy may be a better option for such patients.
“The ability to preserve the blood for several days and still be able to pick up this clinically relevant biomarker is remarkable,” said study author David Miyamoto, MD, PhD, of MGH Cancer Center.
“This is very exciting for clinicians because AR-V7 mRNA can only be detected using CTCs and not with circulating tumor DNA or other cell-free assays.”
The researchers highlighted the universal nature of their blood preservation approach by pointing to its compatibility with the microfluidic CTC-iChip device, which isolates tumor cells by rapid removal of blood cells. The team said this suggests the potential impact of this work extends beyond cancer detection.
“With exciting breakthroughs in immunotherapy, stem cell transplantation, and regenerative medicine—in which peripheral blood is often the source of cells for functional assays or ex vivo expansion—the ability to preserve live cells will greatly ease logistical timelines and reduce the cost of complex cell-based assays,” Dr Wong said.
A new blood stabilization method significantly prolongs the lifespan of blood samples for microfluidic sorting and transcriptome profiling of rare circulating tumor cells (CTCs), according to researchers.
The method involves reducing the storage temperature to 4° C to reversibly slow down cellular processes while also counteracting the platelet activation that can occur because of the low temperature.
The researchers said this work overcomes a significant barrier to the translation of liquid biopsy technologies for precision oncology and other applications.
Keith Wong, PhD, of the Massachusetts General Hospital Center for Engineering in Medicine (MGH-CEM), and his colleagues described this work in Nature Communications.
When isolating CTCs from fresh, unprocessed blood, timing is everything. Even minor changes in the quality of a blood sample—such as the breakdown of red cells, leukocyte activation, or clot formation— can greatly affect cell-sorting mechanisms and the quality of the biomolecules isolated for cancer detection.
According to published studies, factors such as the total number of CTCs in a sample and the number with high-quality RNA decrease by around 50% within the first 4 to 5 hours after the sample is collected.
“At Mass. General, we have the luxury of being so integrated with the clinical team that we can process blood specimens in the lab typically within an hour or 2 after they are drawn,” Dr Wong said.
“But to make these liquid biopsy technologies routine lab tests for the rest of the world, we need ways to keep blood alive for much longer than several hours, since these assays are best performed in central laboratories for reasons of cost-effectiveness and reproducibility.”
With this in mind, Dr Wong and his colleagues set out to preserve blood in its native state with minimal alterations.
“We wanted to slow down the biological clock as much as possible by using hypothermia, but that is not as simple as it sounds,” said study author Shannon Tessier, PhD, also of MGH-CEM.
“Low temperature is a powerful means to decrease metabolism, but a host of unwanted side effects occur at the same time. In some ways, these challenges are similar to those we face in organ preservation, where we have to optimize strategies for a very complex mix of cells.”
To achieve these goals, the researchers first analyzed the effects of hypothermic storage conditions.
The team found that hypothermic storage (4° C) of blood anticoagulated with acid citrate dextrose maintained “cellular morphology, integrity, and surface epitope stability of diverse hematologic cell types” over 72 hours.
However, the researchers also observed platelet activation.
“We are preserving the blood very well, including the coagulation function of platelets,” Dr Wong said. “But, unfortunately, cooling causes profound activation of platelets. Now, we need a targeted approach for platelets so they don’t form nasty clots in the microfluidic blood-sorting device.”
Fortunately, the researchers found that glycoprotein IIb/IIIa inhibitors were able to counter cooling-induced platelet aggregation. And ion chelation treatment with ethylenediaminetetraacetic acid removed activated platelets from leukocytes.
The team said these steps—hypothermic storage, treatment with glycoprotein IIb/IIIa inhibitors, and ion chelation—allowed whole blood preserved for 3 days to be processed as if it were freshly drawn, with very high purity and virtually no loss in CTC numbers.
“The critical achievement here is that the isolated tumor cells contain high-quality RNA that is suitable for demanding molecular assays, such as single-cell qPCR, droplet digital PCR, and RNA sequencing,” Dr Tessier said.
To test their blood preservation method, Dr Tessier and her colleagues used blood specimens from a group of 10 patients with metastatic prostate cancer.
The researchers compared CTC analysis in preserved blood samples and paired fresh samples from the same patients.
There was 92% agreement in the detection of 12 cancer-specific gene transcripts between the fresh and preserved blood samples.
In addition, there was 100% agreement in the detection of a transcript called AR-V7. Recently published studies showed that the presence of AR-V7 mRNA in prostate cancer CTCs predicts resistance to androgen receptor inhibitors, indicating that chemotherapy may be a better option for such patients.
“The ability to preserve the blood for several days and still be able to pick up this clinically relevant biomarker is remarkable,” said study author David Miyamoto, MD, PhD, of MGH Cancer Center.
“This is very exciting for clinicians because AR-V7 mRNA can only be detected using CTCs and not with circulating tumor DNA or other cell-free assays.”
The researchers highlighted the universal nature of their blood preservation approach by pointing to its compatibility with the microfluidic CTC-iChip device, which isolates tumor cells by rapid removal of blood cells. The team said this suggests the potential impact of this work extends beyond cancer detection.
“With exciting breakthroughs in immunotherapy, stem cell transplantation, and regenerative medicine—in which peripheral blood is often the source of cells for functional assays or ex vivo expansion—the ability to preserve live cells will greatly ease logistical timelines and reduce the cost of complex cell-based assays,” Dr Wong said.