User login
Letter to the Editor
Is Hemolysis a Clinical Marker of Propionibacterium acnes Orthopedic Infection or a Phylogenetic Marker?
We read with great interest the study by Nodzo and colleagues in the May 2014 issue of The American Journal of Orthopedics on hemolysis as a clinical marker for Propionibacterium acnes orthopedic infection.1 We agree with the authors that determining if a P acnes culture is a true infection or a contaminant remains a challenge. Although P acnes is described as a commensal bacterium with a low pathogenicity, its involvement has been reported in many clinical entities, especially device-related infections.2P acnes is usually the cause of delayed infections occurring 3 to 24 months or more after prosthesis placement. The rate of P acnes involvement, probably underestimated, is about 10%.3 Although this bacterium was considered to be a contaminant, several virulence factors have been recently identified: putative hemolysins or cytotoxins (CAMP factors, hemolysin III) and enzymes putatively involved in degrading host tissue or molecules (GehA lipase, lysophospholipase, hyaluronate lyase, endoglycoceramidase, etc).4
Interestingly, Nodzo and colleagues revealed that 13 out of 22 P acnes strains were hemolytic and, among them, 10 were considered as definite infections, including 3 with only 1 positive sample. The authors could not identify a statistically significant trend, probably because their study was underpowered due to the size of this case series, as discussed by the authors. Nevertheless, the hemolytic activity of the strains was investigated in the 1980s by adding different concentrations of blood obtained from rabbits, sheep, or humans.5 The hemolytic activity was recorded as positive when a clear, colorless zone around the colonies appeared or weak when slight and incomplete hemolysis under the colonies was found.5 Depending on the erythrocyte origin, differences in the lytic action of hemolysin or cytotoxin may indicate the existence of various enzymes. These enzymes could have different levels of production and provide a distinct hemolytic profile. This hemolytic activity observation could also be correlated to the genetic background of the isolates.
In fact, from a genetic and epidemiological point of view, the sequence analysis of recA gene distinguished 2 distinct lineages of P acnes: types I and II.4 The association of some strains with specific clinical presentations was also demonstrated. Later, McDowell and colleagues6 reported 5 main phylogenetically distinct groups: IA, IB, IC, II, and III. It would have been interesting to know the phylogenetic groups of the strains tested in the study by Nodzo and coauthors, especially as Sampedro and colleagues7 recently reported more phylogenetic groups IA and IB among P acnes strains involved in bone and joint infections. Both of these phylotypes are hemolytic, unlike phylotypes II and III, less often encountered in this clinical entity as reported recently.8 We agree with the authors that hemolytic behavior may be one of the key factors in the variability in the pathogenicity of P acnes strains, suggesting that some strains could be more aggressive than others during deep infection. Another feature is likely the biofilm-production ability of the strains.9,10
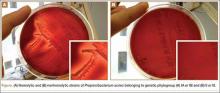
According to our experience, the hemolysis behavior was slightly different depending on which blood agar plates were used to detect hemolytic properties. We have selected 8 isolates or reference ATCC strains from different phylotypes. Each isolate was seeded on 5 different blood agar plates with erythrocyte from various origins (Table). We can confirm that only strains belonging to IA and IB phylotypes were hemolytic, with different behavior as previously reported (Figure).8 Similarly, within IA phylotype strains, the hemolytic property could be different suggesting a difference in the genetic background. However, as the genes encoding all 5 CAMP factors are present in all P acnes groups studied by Valanne and colleagues11 (IA, IB, and II), observed differences reflected different levels of expression rather than missing genes. Moreover, when camp2 or camp4 genes were deleted, the ∆camp2 but not the ∆camp4 mutant exhibited reduced hemolytic activity with sheep erythrocytes, indicating that CAMP factor 2 seems to be the major active cohemolytic factor, but in an IA phylotype P acnes genetic background.12
To conclude, the link between hemolysis and P acnes deep infection remains controversial and complex. The phenotypic differences observed between strains from various types reflect deeper differences in their phylogeny. The hemolytic ability raises the possibility that strains may also display a specific behavior according to their type and variation in their expression of putative virulence factors, including hemolysin, cytotoxin, or lipase. Further studies are clearly needed to better understand the virulence and phylogeny of P acnes strains in order to distinguish contamination from bone infection.
Stéphane Corvec, PharmD, PhD, Jérémy Luchetta, MSc, and Guillaume Ghislain Aubin, PharmD
Nantes University Hospital, Microbiology Laboratory, Nantes, France
Authors’ Response
Corvec and colleagues wrote an interesting summary and make excellent points about the role of hemolysis in Propionibacterium acnes. P acnes upper extremity infection has become an increasingly recognized problem, and determining whether a P acnes culture represents a true infection or a contaminant is still a challenge. We performed this study in hopes of finding an easily usable characteristic of P acnes that would assist the clinician in identifying P acnes strains as true infections rather than contaminants.
Certain pathogenic characteristics of P acnes have been identified, but the clinical implications of this bacterium are still being evaluated. We recognize that the hemolysis phenotype is a characteristic, and may not be the main pathogenic feature, of certain phylotypes of P acnes. It is possible the hemolytic strains in our study were from the IA and IB phylotypes, but, unfortunately, we did not specifically evaluate for phylogeny in our study. This would have correlated well with the work of Sampedro and colleagues,1 which suggested most deep bone and joint infections occur with type IA and IB P acnes phylotypes. Although less common in orthopedic infections, the type II and III phylotypes of P acnes are also capable of causing deep infection, and may not cause a hemolytic reaction on blood agar, which may be why we had some patients classified as a definite infection that did not have a hemolytic strain of P acnes. It is also possible a hemolytic strain may truly be a contaminant, but we did not observe this in our small case series. A larger series may help elucidate this finding, but the majority of truly infected patients in our case series had a hemolytic P acnes phenotype.
The type of blood agar used could have also influenced our results, as noted in the Table in Corvec and colleagues’ letter. We observed the most robust hemolysis on brucella blood agar, and limited hemolysis on CDC (Centers for Disease Control and Prevention) anaerobe blood agar; however, we did not evaluate multiple different blood agar preparations, which could have identified more hemolytic strains.
In our study, the presence of hemolysis was helpful in determining whether or not a true infection existed, but the absence of the hemolytic phenotype did not offer much additional information. The hemolytic phenotype may be a potential marker for those strains that are more aggressive and possibly represent the IA and IB phylotypes, which, as previously stated, are more commonly found in deep bone and joint infections.1 Hemolysis may serve as a surrogate marker for determining these phylotypes since determining phylogeny in a hospital laboratory is burdensome and not possible in most institutions.
In summary, we agree the hemolytic phenotype is commonly observed in certain P acnes phylotypes, and that not all upper extremity orthopedic P acnes infections will have a hemolytic finding. The genetic differences in P acnes strains are complex, and finding a marker of truly pathogenic strains has yet to be established. Larger studies evaluating the clinical outcomes and laboratory findings of patients with and without hemolytic strains of P acnes and evaluating which blood agar is the best at identifying the hemolytic phenotype may be beneficial. Identifying or combining multiple clinical and microbe-specific characteristics may also help guide treatment recommendations when a positive P acnes culture is identified.
Scott R. Nodzo, MD
John K. Crane, MD, PhD
Thomas R. Duquin, MD
Department of Orthopedics
University at Buffalo
Buffalo, NY
Letter to the Editor
1. Nodzo SR, Hohman DW, Crane JK, Duquin TR. Hemolysis as a clinical marker for Propionibacterium acnes orthopedic infection. Am J Orthop. 2014;43(5):E93-E97.
2. Portillo ME, Corvec S, Borens O, Trampuz A. Propionibacterium acnes: an underestimated pathogen in implant-associated infections. BioMed Res Int. 2013;2013:804391.
3. Corvec S, Portillo ME, Pasticci BM, Borens O, Trampuz A. Epidemiology and new developments in the diagnosis of prosthetic joint infection. Int J Artif Organs. 2012;35(10):923-934.
4. Aubin GG, Portillo ME, Trampuz A, Corvec S. Propionibacterium acnes, an emerging pathogen: from acne to implant-infections, from phylotype to resistance. Médecine Mal Infect. 2014;44(6):241-250.
5. Hoeffler U. Enzymatic and hemolytic properties of Propionibacterium acnes and related bacteria. J Clin Microbiol. 1977;6(6):555-558.
6. McDowell A, Perry AL, Lambert PA, Patrick S. A new phylogenetic group of Propionibacterium acnes. J Med Microbiol. 2008;57(Pt 2):218-224.
7. Sampedro MF, Piper KE, McDowell A, et al. Species of Propionibacterium and Propionibacterium acnes phylotypes associated with orthopedic implants. Diagn Microbiol Infect Dis. 2009;64(2):138-145.
8. Lomholt HB, Kilian M. Population genetic analysis of Propionibacterium acnes identifies a subpopulation and epidemic clones associated with acne. PloS One. 2010;5(8):e12277.
9. Furustrand Tafin U, Corvec S, Betrisey B, Zimmerli W, Trampuz A. Role of rifampin against Propionibacterium acnes biofilm in vitro and in an experimental foreign-body infection model. Antimicrob Agents Chemother. 2012;56(4):1885-1891.
10. Holmberg A, Lood R, Mörgelin M, et al. Biofilm formation by Propionibacterium acnes is a characteristic of invasive isolates. Clin Microbiol Infect. 2009;15(8):787-795.
11. Valanne S, McDowell A, Ramage G, et al. CAMP factor homologues in Propionibacterium acnes: a new protein family differentially expressed by types I and II. Microbiol. 2005;151(Pt 5):1369-1379.
12. Sörensen M, Mak TN, Hurwitz R, et al. Mutagenesis of Propionibacterium acnes and analysis of two CAMP factor knock-out mutants. J Microbiol Methods. 2010;83(2):211-216.
Authors' Response Reference
1. Sampedro MF, Piper KE, McDowell A, et al. Species of Propionibacterium and Propionibacterium acnes phylotypes associated with orthopedic implants. Diagn Microbiol Infect Dis. 2009;64(2):138-145.
Letter to the Editor
Is Hemolysis a Clinical Marker of Propionibacterium acnes Orthopedic Infection or a Phylogenetic Marker?
We read with great interest the study by Nodzo and colleagues in the May 2014 issue of The American Journal of Orthopedics on hemolysis as a clinical marker for Propionibacterium acnes orthopedic infection.1 We agree with the authors that determining if a P acnes culture is a true infection or a contaminant remains a challenge. Although P acnes is described as a commensal bacterium with a low pathogenicity, its involvement has been reported in many clinical entities, especially device-related infections.2P acnes is usually the cause of delayed infections occurring 3 to 24 months or more after prosthesis placement. The rate of P acnes involvement, probably underestimated, is about 10%.3 Although this bacterium was considered to be a contaminant, several virulence factors have been recently identified: putative hemolysins or cytotoxins (CAMP factors, hemolysin III) and enzymes putatively involved in degrading host tissue or molecules (GehA lipase, lysophospholipase, hyaluronate lyase, endoglycoceramidase, etc).4
Interestingly, Nodzo and colleagues revealed that 13 out of 22 P acnes strains were hemolytic and, among them, 10 were considered as definite infections, including 3 with only 1 positive sample. The authors could not identify a statistically significant trend, probably because their study was underpowered due to the size of this case series, as discussed by the authors. Nevertheless, the hemolytic activity of the strains was investigated in the 1980s by adding different concentrations of blood obtained from rabbits, sheep, or humans.5 The hemolytic activity was recorded as positive when a clear, colorless zone around the colonies appeared or weak when slight and incomplete hemolysis under the colonies was found.5 Depending on the erythrocyte origin, differences in the lytic action of hemolysin or cytotoxin may indicate the existence of various enzymes. These enzymes could have different levels of production and provide a distinct hemolytic profile. This hemolytic activity observation could also be correlated to the genetic background of the isolates.
In fact, from a genetic and epidemiological point of view, the sequence analysis of recA gene distinguished 2 distinct lineages of P acnes: types I and II.4 The association of some strains with specific clinical presentations was also demonstrated. Later, McDowell and colleagues6 reported 5 main phylogenetically distinct groups: IA, IB, IC, II, and III. It would have been interesting to know the phylogenetic groups of the strains tested in the study by Nodzo and coauthors, especially as Sampedro and colleagues7 recently reported more phylogenetic groups IA and IB among P acnes strains involved in bone and joint infections. Both of these phylotypes are hemolytic, unlike phylotypes II and III, less often encountered in this clinical entity as reported recently.8 We agree with the authors that hemolytic behavior may be one of the key factors in the variability in the pathogenicity of P acnes strains, suggesting that some strains could be more aggressive than others during deep infection. Another feature is likely the biofilm-production ability of the strains.9,10
According to our experience, the hemolysis behavior was slightly different depending on which blood agar plates were used to detect hemolytic properties. We have selected 8 isolates or reference ATCC strains from different phylotypes. Each isolate was seeded on 5 different blood agar plates with erythrocyte from various origins (Table). We can confirm that only strains belonging to IA and IB phylotypes were hemolytic, with different behavior as previously reported (Figure).8 Similarly, within IA phylotype strains, the hemolytic property could be different suggesting a difference in the genetic background. However, as the genes encoding all 5 CAMP factors are present in all P acnes groups studied by Valanne and colleagues11 (IA, IB, and II), observed differences reflected different levels of expression rather than missing genes. Moreover, when camp2 or camp4 genes were deleted, the ∆camp2 but not the ∆camp4 mutant exhibited reduced hemolytic activity with sheep erythrocytes, indicating that CAMP factor 2 seems to be the major active cohemolytic factor, but in an IA phylotype P acnes genetic background.12
To conclude, the link between hemolysis and P acnes deep infection remains controversial and complex. The phenotypic differences observed between strains from various types reflect deeper differences in their phylogeny. The hemolytic ability raises the possibility that strains may also display a specific behavior according to their type and variation in their expression of putative virulence factors, including hemolysin, cytotoxin, or lipase. Further studies are clearly needed to better understand the virulence and phylogeny of P acnes strains in order to distinguish contamination from bone infection.
Stéphane Corvec, PharmD, PhD, Jérémy Luchetta, MSc, and Guillaume Ghislain Aubin, PharmD
Nantes University Hospital, Microbiology Laboratory, Nantes, France
Authors’ Response
Corvec and colleagues wrote an interesting summary and make excellent points about the role of hemolysis in Propionibacterium acnes. P acnes upper extremity infection has become an increasingly recognized problem, and determining whether a P acnes culture represents a true infection or a contaminant is still a challenge. We performed this study in hopes of finding an easily usable characteristic of P acnes that would assist the clinician in identifying P acnes strains as true infections rather than contaminants.
Certain pathogenic characteristics of P acnes have been identified, but the clinical implications of this bacterium are still being evaluated. We recognize that the hemolysis phenotype is a characteristic, and may not be the main pathogenic feature, of certain phylotypes of P acnes. It is possible the hemolytic strains in our study were from the IA and IB phylotypes, but, unfortunately, we did not specifically evaluate for phylogeny in our study. This would have correlated well with the work of Sampedro and colleagues,1 which suggested most deep bone and joint infections occur with type IA and IB P acnes phylotypes. Although less common in orthopedic infections, the type II and III phylotypes of P acnes are also capable of causing deep infection, and may not cause a hemolytic reaction on blood agar, which may be why we had some patients classified as a definite infection that did not have a hemolytic strain of P acnes. It is also possible a hemolytic strain may truly be a contaminant, but we did not observe this in our small case series. A larger series may help elucidate this finding, but the majority of truly infected patients in our case series had a hemolytic P acnes phenotype.
The type of blood agar used could have also influenced our results, as noted in the Table in Corvec and colleagues’ letter. We observed the most robust hemolysis on brucella blood agar, and limited hemolysis on CDC (Centers for Disease Control and Prevention) anaerobe blood agar; however, we did not evaluate multiple different blood agar preparations, which could have identified more hemolytic strains.
In our study, the presence of hemolysis was helpful in determining whether or not a true infection existed, but the absence of the hemolytic phenotype did not offer much additional information. The hemolytic phenotype may be a potential marker for those strains that are more aggressive and possibly represent the IA and IB phylotypes, which, as previously stated, are more commonly found in deep bone and joint infections.1 Hemolysis may serve as a surrogate marker for determining these phylotypes since determining phylogeny in a hospital laboratory is burdensome and not possible in most institutions.
In summary, we agree the hemolytic phenotype is commonly observed in certain P acnes phylotypes, and that not all upper extremity orthopedic P acnes infections will have a hemolytic finding. The genetic differences in P acnes strains are complex, and finding a marker of truly pathogenic strains has yet to be established. Larger studies evaluating the clinical outcomes and laboratory findings of patients with and without hemolytic strains of P acnes and evaluating which blood agar is the best at identifying the hemolytic phenotype may be beneficial. Identifying or combining multiple clinical and microbe-specific characteristics may also help guide treatment recommendations when a positive P acnes culture is identified.
Scott R. Nodzo, MD
John K. Crane, MD, PhD
Thomas R. Duquin, MD
Department of Orthopedics
University at Buffalo
Buffalo, NY
Letter to the Editor
Is Hemolysis a Clinical Marker of Propionibacterium acnes Orthopedic Infection or a Phylogenetic Marker?
We read with great interest the study by Nodzo and colleagues in the May 2014 issue of The American Journal of Orthopedics on hemolysis as a clinical marker for Propionibacterium acnes orthopedic infection.1 We agree with the authors that determining if a P acnes culture is a true infection or a contaminant remains a challenge. Although P acnes is described as a commensal bacterium with a low pathogenicity, its involvement has been reported in many clinical entities, especially device-related infections.2P acnes is usually the cause of delayed infections occurring 3 to 24 months or more after prosthesis placement. The rate of P acnes involvement, probably underestimated, is about 10%.3 Although this bacterium was considered to be a contaminant, several virulence factors have been recently identified: putative hemolysins or cytotoxins (CAMP factors, hemolysin III) and enzymes putatively involved in degrading host tissue or molecules (GehA lipase, lysophospholipase, hyaluronate lyase, endoglycoceramidase, etc).4
Interestingly, Nodzo and colleagues revealed that 13 out of 22 P acnes strains were hemolytic and, among them, 10 were considered as definite infections, including 3 with only 1 positive sample. The authors could not identify a statistically significant trend, probably because their study was underpowered due to the size of this case series, as discussed by the authors. Nevertheless, the hemolytic activity of the strains was investigated in the 1980s by adding different concentrations of blood obtained from rabbits, sheep, or humans.5 The hemolytic activity was recorded as positive when a clear, colorless zone around the colonies appeared or weak when slight and incomplete hemolysis under the colonies was found.5 Depending on the erythrocyte origin, differences in the lytic action of hemolysin or cytotoxin may indicate the existence of various enzymes. These enzymes could have different levels of production and provide a distinct hemolytic profile. This hemolytic activity observation could also be correlated to the genetic background of the isolates.
In fact, from a genetic and epidemiological point of view, the sequence analysis of recA gene distinguished 2 distinct lineages of P acnes: types I and II.4 The association of some strains with specific clinical presentations was also demonstrated. Later, McDowell and colleagues6 reported 5 main phylogenetically distinct groups: IA, IB, IC, II, and III. It would have been interesting to know the phylogenetic groups of the strains tested in the study by Nodzo and coauthors, especially as Sampedro and colleagues7 recently reported more phylogenetic groups IA and IB among P acnes strains involved in bone and joint infections. Both of these phylotypes are hemolytic, unlike phylotypes II and III, less often encountered in this clinical entity as reported recently.8 We agree with the authors that hemolytic behavior may be one of the key factors in the variability in the pathogenicity of P acnes strains, suggesting that some strains could be more aggressive than others during deep infection. Another feature is likely the biofilm-production ability of the strains.9,10
According to our experience, the hemolysis behavior was slightly different depending on which blood agar plates were used to detect hemolytic properties. We have selected 8 isolates or reference ATCC strains from different phylotypes. Each isolate was seeded on 5 different blood agar plates with erythrocyte from various origins (Table). We can confirm that only strains belonging to IA and IB phylotypes were hemolytic, with different behavior as previously reported (Figure).8 Similarly, within IA phylotype strains, the hemolytic property could be different suggesting a difference in the genetic background. However, as the genes encoding all 5 CAMP factors are present in all P acnes groups studied by Valanne and colleagues11 (IA, IB, and II), observed differences reflected different levels of expression rather than missing genes. Moreover, when camp2 or camp4 genes were deleted, the ∆camp2 but not the ∆camp4 mutant exhibited reduced hemolytic activity with sheep erythrocytes, indicating that CAMP factor 2 seems to be the major active cohemolytic factor, but in an IA phylotype P acnes genetic background.12
To conclude, the link between hemolysis and P acnes deep infection remains controversial and complex. The phenotypic differences observed between strains from various types reflect deeper differences in their phylogeny. The hemolytic ability raises the possibility that strains may also display a specific behavior according to their type and variation in their expression of putative virulence factors, including hemolysin, cytotoxin, or lipase. Further studies are clearly needed to better understand the virulence and phylogeny of P acnes strains in order to distinguish contamination from bone infection.
Stéphane Corvec, PharmD, PhD, Jérémy Luchetta, MSc, and Guillaume Ghislain Aubin, PharmD
Nantes University Hospital, Microbiology Laboratory, Nantes, France
Authors’ Response
Corvec and colleagues wrote an interesting summary and make excellent points about the role of hemolysis in Propionibacterium acnes. P acnes upper extremity infection has become an increasingly recognized problem, and determining whether a P acnes culture represents a true infection or a contaminant is still a challenge. We performed this study in hopes of finding an easily usable characteristic of P acnes that would assist the clinician in identifying P acnes strains as true infections rather than contaminants.
Certain pathogenic characteristics of P acnes have been identified, but the clinical implications of this bacterium are still being evaluated. We recognize that the hemolysis phenotype is a characteristic, and may not be the main pathogenic feature, of certain phylotypes of P acnes. It is possible the hemolytic strains in our study were from the IA and IB phylotypes, but, unfortunately, we did not specifically evaluate for phylogeny in our study. This would have correlated well with the work of Sampedro and colleagues,1 which suggested most deep bone and joint infections occur with type IA and IB P acnes phylotypes. Although less common in orthopedic infections, the type II and III phylotypes of P acnes are also capable of causing deep infection, and may not cause a hemolytic reaction on blood agar, which may be why we had some patients classified as a definite infection that did not have a hemolytic strain of P acnes. It is also possible a hemolytic strain may truly be a contaminant, but we did not observe this in our small case series. A larger series may help elucidate this finding, but the majority of truly infected patients in our case series had a hemolytic P acnes phenotype.
The type of blood agar used could have also influenced our results, as noted in the Table in Corvec and colleagues’ letter. We observed the most robust hemolysis on brucella blood agar, and limited hemolysis on CDC (Centers for Disease Control and Prevention) anaerobe blood agar; however, we did not evaluate multiple different blood agar preparations, which could have identified more hemolytic strains.
In our study, the presence of hemolysis was helpful in determining whether or not a true infection existed, but the absence of the hemolytic phenotype did not offer much additional information. The hemolytic phenotype may be a potential marker for those strains that are more aggressive and possibly represent the IA and IB phylotypes, which, as previously stated, are more commonly found in deep bone and joint infections.1 Hemolysis may serve as a surrogate marker for determining these phylotypes since determining phylogeny in a hospital laboratory is burdensome and not possible in most institutions.
In summary, we agree the hemolytic phenotype is commonly observed in certain P acnes phylotypes, and that not all upper extremity orthopedic P acnes infections will have a hemolytic finding. The genetic differences in P acnes strains are complex, and finding a marker of truly pathogenic strains has yet to be established. Larger studies evaluating the clinical outcomes and laboratory findings of patients with and without hemolytic strains of P acnes and evaluating which blood agar is the best at identifying the hemolytic phenotype may be beneficial. Identifying or combining multiple clinical and microbe-specific characteristics may also help guide treatment recommendations when a positive P acnes culture is identified.
Scott R. Nodzo, MD
John K. Crane, MD, PhD
Thomas R. Duquin, MD
Department of Orthopedics
University at Buffalo
Buffalo, NY
Letter to the Editor
1. Nodzo SR, Hohman DW, Crane JK, Duquin TR. Hemolysis as a clinical marker for Propionibacterium acnes orthopedic infection. Am J Orthop. 2014;43(5):E93-E97.
2. Portillo ME, Corvec S, Borens O, Trampuz A. Propionibacterium acnes: an underestimated pathogen in implant-associated infections. BioMed Res Int. 2013;2013:804391.
3. Corvec S, Portillo ME, Pasticci BM, Borens O, Trampuz A. Epidemiology and new developments in the diagnosis of prosthetic joint infection. Int J Artif Organs. 2012;35(10):923-934.
4. Aubin GG, Portillo ME, Trampuz A, Corvec S. Propionibacterium acnes, an emerging pathogen: from acne to implant-infections, from phylotype to resistance. Médecine Mal Infect. 2014;44(6):241-250.
5. Hoeffler U. Enzymatic and hemolytic properties of Propionibacterium acnes and related bacteria. J Clin Microbiol. 1977;6(6):555-558.
6. McDowell A, Perry AL, Lambert PA, Patrick S. A new phylogenetic group of Propionibacterium acnes. J Med Microbiol. 2008;57(Pt 2):218-224.
7. Sampedro MF, Piper KE, McDowell A, et al. Species of Propionibacterium and Propionibacterium acnes phylotypes associated with orthopedic implants. Diagn Microbiol Infect Dis. 2009;64(2):138-145.
8. Lomholt HB, Kilian M. Population genetic analysis of Propionibacterium acnes identifies a subpopulation and epidemic clones associated with acne. PloS One. 2010;5(8):e12277.
9. Furustrand Tafin U, Corvec S, Betrisey B, Zimmerli W, Trampuz A. Role of rifampin against Propionibacterium acnes biofilm in vitro and in an experimental foreign-body infection model. Antimicrob Agents Chemother. 2012;56(4):1885-1891.
10. Holmberg A, Lood R, Mörgelin M, et al. Biofilm formation by Propionibacterium acnes is a characteristic of invasive isolates. Clin Microbiol Infect. 2009;15(8):787-795.
11. Valanne S, McDowell A, Ramage G, et al. CAMP factor homologues in Propionibacterium acnes: a new protein family differentially expressed by types I and II. Microbiol. 2005;151(Pt 5):1369-1379.
12. Sörensen M, Mak TN, Hurwitz R, et al. Mutagenesis of Propionibacterium acnes and analysis of two CAMP factor knock-out mutants. J Microbiol Methods. 2010;83(2):211-216.
Authors' Response Reference
1. Sampedro MF, Piper KE, McDowell A, et al. Species of Propionibacterium and Propionibacterium acnes phylotypes associated with orthopedic implants. Diagn Microbiol Infect Dis. 2009;64(2):138-145.
Letter to the Editor
1. Nodzo SR, Hohman DW, Crane JK, Duquin TR. Hemolysis as a clinical marker for Propionibacterium acnes orthopedic infection. Am J Orthop. 2014;43(5):E93-E97.
2. Portillo ME, Corvec S, Borens O, Trampuz A. Propionibacterium acnes: an underestimated pathogen in implant-associated infections. BioMed Res Int. 2013;2013:804391.
3. Corvec S, Portillo ME, Pasticci BM, Borens O, Trampuz A. Epidemiology and new developments in the diagnosis of prosthetic joint infection. Int J Artif Organs. 2012;35(10):923-934.
4. Aubin GG, Portillo ME, Trampuz A, Corvec S. Propionibacterium acnes, an emerging pathogen: from acne to implant-infections, from phylotype to resistance. Médecine Mal Infect. 2014;44(6):241-250.
5. Hoeffler U. Enzymatic and hemolytic properties of Propionibacterium acnes and related bacteria. J Clin Microbiol. 1977;6(6):555-558.
6. McDowell A, Perry AL, Lambert PA, Patrick S. A new phylogenetic group of Propionibacterium acnes. J Med Microbiol. 2008;57(Pt 2):218-224.
7. Sampedro MF, Piper KE, McDowell A, et al. Species of Propionibacterium and Propionibacterium acnes phylotypes associated with orthopedic implants. Diagn Microbiol Infect Dis. 2009;64(2):138-145.
8. Lomholt HB, Kilian M. Population genetic analysis of Propionibacterium acnes identifies a subpopulation and epidemic clones associated with acne. PloS One. 2010;5(8):e12277.
9. Furustrand Tafin U, Corvec S, Betrisey B, Zimmerli W, Trampuz A. Role of rifampin against Propionibacterium acnes biofilm in vitro and in an experimental foreign-body infection model. Antimicrob Agents Chemother. 2012;56(4):1885-1891.
10. Holmberg A, Lood R, Mörgelin M, et al. Biofilm formation by Propionibacterium acnes is a characteristic of invasive isolates. Clin Microbiol Infect. 2009;15(8):787-795.
11. Valanne S, McDowell A, Ramage G, et al. CAMP factor homologues in Propionibacterium acnes: a new protein family differentially expressed by types I and II. Microbiol. 2005;151(Pt 5):1369-1379.
12. Sörensen M, Mak TN, Hurwitz R, et al. Mutagenesis of Propionibacterium acnes and analysis of two CAMP factor knock-out mutants. J Microbiol Methods. 2010;83(2):211-216.
Authors' Response Reference
1. Sampedro MF, Piper KE, McDowell A, et al. Species of Propionibacterium and Propionibacterium acnes phylotypes associated with orthopedic implants. Diagn Microbiol Infect Dis. 2009;64(2):138-145.