User login
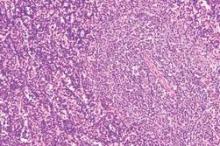
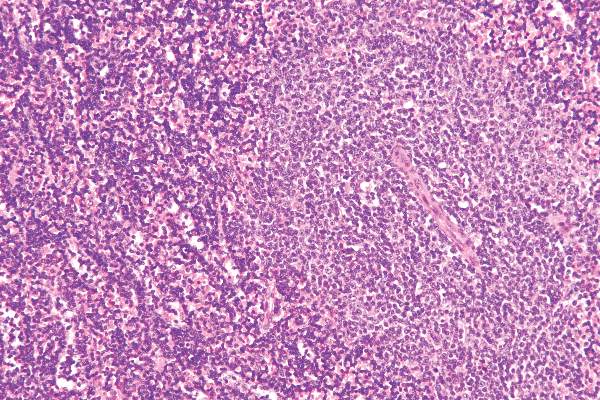
Researchers have devised a simple and reproducible method of tracking the cellular origin of chronic lymphocytic leukemia (CLL) by applying five epigenetic biomarkers. By using this strategy, CLL patients can be categorized into three epigenetic subgroups with differential clinicobiologic features and outcomes – naive B-cell-like, intermediate, and memory B-cell-like CLL, according to a paper published in Leukemia.
Being able to identify CLL patients early on who are destined to progress would greatly help their clinical management, said Dr. Ana C. Queirós of the University of Barcelona and colleagues (Leukemia. 2015;29:598-605.
“We believe that the most relevant information obtained by the five epigenetic biomarkers is to classify CLL patients based on the putative cell of origin of the disease rather than being a mere additional prognostic biomarker,” the investigators wrote. “The recent advance in the genetics and cellular biology of CLL, including the present epigenetic classification, could result in the use of targeted therapies for specific subgroups of patients.”
In previous research, the authors identified the presence of three subgroups of CLL with different clinicobiologic features, and in this study they hypothesized that DNA methylation patterns associated with normal B cells could be used to classify CLL into three novel subgroups.
To test their hypothesis and to develop a clinically useful strategy, they identified five epigenetic biomarkers and established new quantitative DNA methylation assays, applying them to two independent series of CLL patients of different geographical origin.
The first epigenetic classification was determined in an initial cohort of 211 CLL patients and then validated in a series of 97 additional CLL patients.
To test the stability of these markers over time and after treatment, two or three sequential samples were analyzed from 27 CLL patients with a median difference between samples of 59 months (range, 5-114). In addition, specimens from 13 patients, from before and after treatment, also were analyzed.
In the initial 211 patients, the three subgroups had different levels of immunoglobulin heavy-chain locus (IGHV) mutation (P <.001) and VH gene usage (P <.03). There also were different clinical features and outcomes in regard to their time to first treatment and overall survival (P <.001), Dr. Queirós and associates reported.
After a Cox multivariate analysis, the final model showed that the epigenetic signature related to the cellular origin of CLL was the most important variable in predicting time to first treatment. Other important variables in the model were Binet stage, CD38 expression, LDH levels, and SF3B1 mutations.
The study was funded by the Spanish Ministry of Economy and Competitiveness (MINECO) through the Instituto de Salud Carlos III (ISCIII) and the Red Temática de Investigación del Cáncer (RTICC) of the ISCIII and project SAF2009-08663, the UK Medical Research Council, and the European Union’s Seventh Framework Programme through the Blueprint Consortium. The authors declared no conflicts of interest.
Researchers have devised a simple and reproducible method of tracking the cellular origin of chronic lymphocytic leukemia (CLL) by applying five epigenetic biomarkers. By using this strategy, CLL patients can be categorized into three epigenetic subgroups with differential clinicobiologic features and outcomes – naive B-cell-like, intermediate, and memory B-cell-like CLL, according to a paper published in Leukemia.
Being able to identify CLL patients early on who are destined to progress would greatly help their clinical management, said Dr. Ana C. Queirós of the University of Barcelona and colleagues (Leukemia. 2015;29:598-605.
“We believe that the most relevant information obtained by the five epigenetic biomarkers is to classify CLL patients based on the putative cell of origin of the disease rather than being a mere additional prognostic biomarker,” the investigators wrote. “The recent advance in the genetics and cellular biology of CLL, including the present epigenetic classification, could result in the use of targeted therapies for specific subgroups of patients.”
In previous research, the authors identified the presence of three subgroups of CLL with different clinicobiologic features, and in this study they hypothesized that DNA methylation patterns associated with normal B cells could be used to classify CLL into three novel subgroups.
To test their hypothesis and to develop a clinically useful strategy, they identified five epigenetic biomarkers and established new quantitative DNA methylation assays, applying them to two independent series of CLL patients of different geographical origin.
The first epigenetic classification was determined in an initial cohort of 211 CLL patients and then validated in a series of 97 additional CLL patients.
To test the stability of these markers over time and after treatment, two or three sequential samples were analyzed from 27 CLL patients with a median difference between samples of 59 months (range, 5-114). In addition, specimens from 13 patients, from before and after treatment, also were analyzed.
In the initial 211 patients, the three subgroups had different levels of immunoglobulin heavy-chain locus (IGHV) mutation (P <.001) and VH gene usage (P <.03). There also were different clinical features and outcomes in regard to their time to first treatment and overall survival (P <.001), Dr. Queirós and associates reported.
After a Cox multivariate analysis, the final model showed that the epigenetic signature related to the cellular origin of CLL was the most important variable in predicting time to first treatment. Other important variables in the model were Binet stage, CD38 expression, LDH levels, and SF3B1 mutations.
The study was funded by the Spanish Ministry of Economy and Competitiveness (MINECO) through the Instituto de Salud Carlos III (ISCIII) and the Red Temática de Investigación del Cáncer (RTICC) of the ISCIII and project SAF2009-08663, the UK Medical Research Council, and the European Union’s Seventh Framework Programme through the Blueprint Consortium. The authors declared no conflicts of interest.
Researchers have devised a simple and reproducible method of tracking the cellular origin of chronic lymphocytic leukemia (CLL) by applying five epigenetic biomarkers. By using this strategy, CLL patients can be categorized into three epigenetic subgroups with differential clinicobiologic features and outcomes – naive B-cell-like, intermediate, and memory B-cell-like CLL, according to a paper published in Leukemia.
Being able to identify CLL patients early on who are destined to progress would greatly help their clinical management, said Dr. Ana C. Queirós of the University of Barcelona and colleagues (Leukemia. 2015;29:598-605.
“We believe that the most relevant information obtained by the five epigenetic biomarkers is to classify CLL patients based on the putative cell of origin of the disease rather than being a mere additional prognostic biomarker,” the investigators wrote. “The recent advance in the genetics and cellular biology of CLL, including the present epigenetic classification, could result in the use of targeted therapies for specific subgroups of patients.”
In previous research, the authors identified the presence of three subgroups of CLL with different clinicobiologic features, and in this study they hypothesized that DNA methylation patterns associated with normal B cells could be used to classify CLL into three novel subgroups.
To test their hypothesis and to develop a clinically useful strategy, they identified five epigenetic biomarkers and established new quantitative DNA methylation assays, applying them to two independent series of CLL patients of different geographical origin.
The first epigenetic classification was determined in an initial cohort of 211 CLL patients and then validated in a series of 97 additional CLL patients.
To test the stability of these markers over time and after treatment, two or three sequential samples were analyzed from 27 CLL patients with a median difference between samples of 59 months (range, 5-114). In addition, specimens from 13 patients, from before and after treatment, also were analyzed.
In the initial 211 patients, the three subgroups had different levels of immunoglobulin heavy-chain locus (IGHV) mutation (P <.001) and VH gene usage (P <.03). There also were different clinical features and outcomes in regard to their time to first treatment and overall survival (P <.001), Dr. Queirós and associates reported.
After a Cox multivariate analysis, the final model showed that the epigenetic signature related to the cellular origin of CLL was the most important variable in predicting time to first treatment. Other important variables in the model were Binet stage, CD38 expression, LDH levels, and SF3B1 mutations.
The study was funded by the Spanish Ministry of Economy and Competitiveness (MINECO) through the Instituto de Salud Carlos III (ISCIII) and the Red Temática de Investigación del Cáncer (RTICC) of the ISCIII and project SAF2009-08663, the UK Medical Research Council, and the European Union’s Seventh Framework Programme through the Blueprint Consortium. The authors declared no conflicts of interest.
FROM LEUKEMIA
Key clinical point: A new strategy allows CLL patients to be categorized into three epigenetic subgroups with differential clinicobiologic features and outcomes.
Major finding: Epigenetic classification was the strongest predictor of time to treatment (P <.001), along with Binet stage (P <.001); these findings were corroborated in a validation series (n = 97).
Data source: A prediction model formulated using five epigenetic biomarkers that was able to classify CLL patients accurately into the three subgroups.
Disclosures: The study was funded by the Spanish Ministry of Economy and Competitiveness (MINECO) through the Instituto de Salud Carlos III (ISCIII) and the Red Temática de Investigación del Cáncer (RTICC) of the ISCIII and project SAF2009-08663, the UK Medical Research Council, and the European Union’s Seventh Framework Programme through the Blueprint Consortium. The authors declared no conflicts of interest.